 |
PDBsum entry 1j2c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1j2c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.14.18
- heme oxygenase (biliverdin-producing).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
heme b + 3 reduced [NADPH--hemoprotein reductase] + 3 O2 = biliverdin IXalpha + CO + Fe2+ + 3 oxidized [NADPH--hemoprotein reductase] + 3 H2O + H+
|
 |
 |
 |
 |
 |
heme b
Bound ligand (Het Group name = )
matches with 77.78% similarity
|
+
|
3
×
reduced [NADPH--hemoprotein reductase]
|
+
|
 3
×
O2
3
×
O2
|
=
|
biliverdin IXalpha
|
+
|
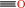 CO
CO
|
+
|
Fe(2+)
|
+
|
3
×
oxidized [NADPH--hemoprotein reductase]
|
+
|
3
×
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:32352-32358
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of rat heme oxygenase-1 in complex with biliverdin-iron chelate. Conformational change of the distal helix during the heme cleavage reaction.
|
|
M.Sugishima,
H.Sakamoto,
Y.Higashimoto,
M.Noguchi,
K.Fukuyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of rat heme oxygenase-1 in complex with biliverdin-iron
chelate (biliverdin(Fe)-HO-1), the immediate precursor of the final product,
biliverdin, has been determined at a 2.4-A resolution. The electron density in
the heme pocket clearly showed that the tetrapyrrole ring of heme is cleaved at
the alpha-meso edge. Like the heme bound to HO-1, biliverdin-iron chelate is
located between the distal and proximal helices, but its accommodation state
seems to be less stable in light of the disordering of the solvent-exposed
propionate and vinyl groups. The middle of the distal helix is shifted away from
the center of the active site in biliverdin(Fe)-HO-1, increasing the size of the
heme pocket. The hydrogen-bonding interaction between Glu-29 and Gln-38,
considered to restrain the orientation of the proximal helix in the heme-HO-1
complex, was lost in biliverdin(Fe)-HO-1, leading to relaxation of the helix.
Biliverdin has a distorted helical conformation; the lactam oxygen atom of its
pyrrole ring-A interacted with Asp-140 through a hydrogen-bonding solvent
network. Because of the absence of a distal water ligand, the iron atom is
five-coordinated with His-25 and four pyrrole nitrogen atoms. The coordination
geometry deviates considerably from a square pyramid, suggesting that the iron
may be readily dissociated. We speculate that the opened conformation of the
heme pocket facilitates sequential product release, first iron then biliverdin,
and that because of biliverdin's increased flexibility, iron release triggers
its slow dissociation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
FIG. 2. Comparison of the heme pocket structures of
biliverdin(Fe)-HO-1 and N[3]^--heme-HO-1. Ball and stick models
of the heme pockets of biliverdin(Fe)-HO-1 (left) and
N[3]^--heme-HO-1 (right; Protein Data Bank code 1IVJ [PDB]
). For clarity, in the F helix only main-chain atoms are shown.
Dashed lines indicate hydrogen-bonds that stabilize the two
conformers. Numerals show distances between atoms in Angstroms.
|
 |
Figure 6.
FIG. 6. Comparison of the distal hydrogen-bond networks in
biliverdin(Fe)-HO-1 (green) and N[3]^--heme-HO-1 (light blue;
Protein Data Bank code 1IVJ [PDB]
). W1 through W5 indicate the water molecules in
N[3]^--heme-HO-1 (17). Dashed lines indicate hydrogen-bonds
(thin lines, in N[3]^--heme-HO-1; bold lines, in
biliverdin(Fe)-HO-1).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
32352-32358)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Hagiwara,
M.Sugishima,
H.Khawn,
H.Kinoshita,
K.Inomata,
L.Shang,
J.C.Lagarias,
Y.Takahashi,
and
K.Fukuyama
(2010).
Structural insights into vinyl reduction regiospecificity of phycocyanobilin:ferredoxin oxidoreductase (PcyA).
|
| |
J Biol Chem,
285,
1000-1007.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.H.Ma,
Y.Liu,
X.Zhang,
T.Yoshida,
and
G.N.La Mar
(2009).
1H NMR study of the effect of variable ligand on heme oxygenase electronic and molecular structure.
|
| |
J Inorg Biochem,
103,
10-19.
|
 |
|
|
|
|
 |
W.J.Huber Iii,
B.A.Scruggs,
and
W.L.Backes
(2009).
C-Terminal membrane spanning region of human heme oxygenase-1 mediates a time-dependent complex formation with cytochrome P450 reductase.
|
| |
Biochemistry,
48,
190-197.
|
 |
|
|
|
|
 |
M.Bröring,
S.Köhler,
S.Link,
O.Burghaus,
C.Pietzonka,
H.Kelm,
and
H.J.Krüger
(2008).
Iron chelates of 2,2'-bidipyrrin: stable analogues of the labile iron bilins.
|
| |
Chemistry,
14,
4006-4016.
|
 |
|
|
|
|
 |
P.R.Jamaat,
N.Safari,
M.Ghiasi,
S.S.Naghavi,
and
M.Zahedi
(2008).
Noninnocent effect of axial ligand on the heme degradation process: a theoretical approach to hydrolysis pathway of verdoheme to biliverdin.
|
| |
J Biol Inorg Chem,
13,
121-132.
|
 |
|
|
|
|
 |
Y.Higashimoto,
M.Sugishima,
H.Sato,
H.Sakamoto,
K.Fukuyama,
G.Palmer,
and
M.Noguchi
(2008).
Mass spectrometric identification of lysine residues of heme oxygenase-1 that are involved in its interaction with NADPH-cytochrome P450 reductase.
|
| |
Biochem Biophys Res Commun,
367,
852-858.
|
 |
|
|
|
|
 |
C.M.Bianchetti,
L.Yi,
S.W.Ragsdale,
and
G.N.Phillips
(2007).
Comparison of apo- and heme-bound crystal structures of a truncated human heme oxygenase-2.
|
| |
J Biol Chem,
282,
37624-37631.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Unno,
T.Matsui,
and
M.Ikeda-Saito
(2007).
Structure and catalytic mechanism of heme oxygenase.
|
| |
Nat Prod Rep,
24,
553-570.
|
 |
|
|
|
|
 |
L.Lad,
A.Koshkin,
P.R.de Montellano,
and
T.L.Poulos
(2005).
Crystal structures of the G139A, G139A-NO and G143H mutants of human heme oxygenase-1. A finely tuned hydrogen-bonding network controls oxygenase versus peroxidase activity.
|
| |
J Biol Inorg Chem,
10,
138-146.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Sugishima,
C.T.Migita,
X.Zhang,
T.Yoshida,
and
K.Fukuyama
(2004).
Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC 6803 in complex with heme.
|
| |
Eur J Biochem,
271,
4517-4525.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

