 |
PDBsum entry 1ipd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ipd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.85
- 3-isopropylmalate dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-isopropylmalate + NAD+ = 4-methyl-2-oxopentanoate + CO2 + NADH
|
 |
 |
 |
 |
 |
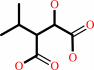 (2R,3S)-3-isopropylmalate
(2R,3S)-3-isopropylmalate
|
+
|
 NAD(+)
NAD(+)
|
=
|
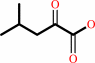 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 CO2
CO2
|
+
|
 NADH
NADH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
222:725-738
(1991)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
|
|
K.Imada,
M.Sato,
N.Tanaka,
Y.Katsube,
Y.Matsuura,
T.Oshima.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The three-dimensional structure of the highly thermostable 3-isopropylmalate
dehydrogenase (IPMDH) from Thermus thermophilus has been determined by the
multiple isomorphous replacement method and refined to 2.2 A resolution. The
final R-factor is 0.185 for 20,307 reflections. The crystal asymmetric unit has
one subunit consisting of 345 amino acid residues. The polypeptide chain of this
subunit is folded into two domains (first and second domains) with parallel
alpha/beta motifs. The domains are similar in their conformations and folding
topologies, but differ from those of the NAD-binding domains of such well-known
enzymes as the alcohol and lactate dehydrogenases. A beta-strand that is a part
of the long arm-like polypeptide protruding from the second domain comes into
contact with another subunit and contributes to the formation of an isologous
dimer with a crystallographic 2-fold symmetry. Close subunit contacts are also
present at two alpha-helices in the second domain. These helices strongly
interact hydrophobically with the corresponding helices of the other subunit to
form a hydrophobic core at the center of the dimer. Two large pockets that exist
between the first domain of one subunit and the second domain of the other
include the amino acid residues responsible for substrate binding. These results
indicate that the dimeric form is essential for the IPMDH to express enzymatic
activity and that the close subunit contact at the hydrophobic core is important
for the thermal stability of the enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Ã.‰.Gráczer,
A.Merli,
R.K.Singh,
M.Karuppasamy,
P.Závodszky,
M.S.Weiss,
and
M.Vas
(2011).
Atomic level description of the domain closure in a dimeric enzyme: thermus thermophilus 3-isopropylmalate dehydrogenase.
|
| |
Mol Biosyst,
7,
1646-1659.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Lunzer,
G.B.Golding,
and
A.M.Dean
(2010).
Pervasive cryptic epistasis in molecular evolution.
|
| |
PLoS Genet,
6,
e1001162.
|
 |
|
|
|
|
 |
I.Hajdú,
A.Szilágyi,
J.Kardos,
and
P.Závodszky
(2009).
A link between hinge-bending domain motions and the temperature dependence of catalysis in 3-isopropylmalate dehydrogenase.
|
| |
Biophys J,
96,
5003-5012.
|
 |
|
|
|
|
 |
K.Homma,
and
H.Moriyama
(2009).
Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
1030-1034.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Kasahara,
T.Sato,
H.Tamegai,
and
C.Kato
(2009).
Piezo-adapted 3-isopropylmalate dehydrogenase of the obligate piezophile Shewanella benthica DB21MT-2 isolated from the 11,000-m depth of the Mariana Trench.
|
| |
Biosci Biotechnol Biochem,
73,
2541-2543.
|
 |
|
|
|
|
 |
A.B.Taylor,
G.Hu,
P.J.Hart,
and
L.McAlister-Henn
(2008).
Allosteric motions in structures of yeast NAD+-specific isocitrate dehydrogenase.
|
| |
J Biol Chem,
283,
10872-10880.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Imada,
T.Tamura,
R.Takenaka,
I.Kobayashi,
K.Namba,
and
K.Inagaki
(2008).
Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity.
|
| |
Proteins,
70,
63-71.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Aoshima,
and
Y.Igarashi
(2008).
Nondecarboxylating and decarboxylating isocitrate dehydrogenases: oxalosuccinate reductase as an ancestral form of isocitrate dehydrogenase.
|
| |
J Bacteriol,
190,
2050-2055.
|
 |
|
|
|
|
 |
M.Sasaki,
M.Uno,
S.Akanuma,
and
A.Yamagishi
(2008).
Random mutagenesis improves the low-temperature activity of the tetrameric 3-isopropylmalate dehydrogenase from the hyperthermophile Sulfolobus tokodaii.
|
| |
Protein Eng Des Sel,
21,
721-727.
|
 |
|
|
|
|
 |
R.Stokke,
D.Madern,
A.E.Fedøy,
S.Karlsen,
N.K.Birkeland,
and
I.H.Steen
(2007).
Biochemical characterization of isocitrate dehydrogenase from Methylococcus capsulatus reveals a unique NAD+-dependent homotetrameric enzyme.
|
| |
Arch Microbiol,
187,
361-370.
|
 |
|
|
|
|
 |
J.A.McCourt,
and
R.G.Duggleby
(2006).
Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids.
|
| |
Amino Acids,
31,
173-210.
|
 |
|
|
|
|
 |
M.Karlström,
I.H.Steen,
D.Madern,
A.E.Fedöy,
N.K.Birkeland,
and
R.Ladenstein
(2006).
The crystal structure of a hyperthermostable subfamily II isocitrate dehydrogenase from Thermotoga maritima.
|
| |
FEBS J,
273,
2851-2868.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.V.Kalinina,
and
M.S.Gelfand
(2006).
Amino acid residues that determine functional specificity of NADP- and NAD-dependent isocitrate and isopropylmalate dehydrogenases.
|
| |
Proteins,
64,
1001-1009.
|
 |
|
|
|
|
 |
A.Rodríguez-Arnedo,
M.Camacho,
F.Llorca,
and
M.J.Bonete
(2005).
Complete reversal of coenzyme specificity of isocitrate dehydrogenase from Haloferax volcanii.
|
| |
Protein J,
24,
259-266.
|
 |
|
|
|
|
 |
G.Hu,
A.B.Taylor,
L.McAlister-Henn,
and
P.J.Hart
(2005).
Crystallization and preliminary X-ray crystallographic analysis of yeast NAD+-specific isocitrate dehydrogenase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
486-488.
|
 |
|
|
|
|
 |
H.Iwabata,
K.Watanabe,
T.Ohkuri,
S.Yokobori,
and
A.Yamagishi
(2005).
Thermostability of ancestral mutants of Caldococcus noboribetus isocitrate dehydrogenase.
|
| |
FEMS Microbiol Lett,
243,
393-398.
|
 |
|
|
|
|
 |
H.Turakainen,
and
M.Korhola
(2005).
Cloning, sequencing and application of the LEU2 gene from the sour dough yeast Candida milleri.
|
| |
Yeast,
22,
805-812.
|
 |
|
|
|
|
 |
J.Miyazaki,
K.Asada,
S.Fushinobu,
T.Kuzuyama,
and
M.Nishiyama
(2005).
Crystal structure of tetrameric homoisocitrate dehydrogenase from an extreme thermophile, Thermus thermophilus: involvement of hydrophobic dimer-dimer interaction in extremely high thermotolerance.
|
| |
J Bacteriol,
187,
6779-6788.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Okochi,
H.Matsuzaki,
T.Nomura,
N.Ishii,
and
M.Yohda
(2005).
Molecular characterization of the group II chaperonin from the hyperthermophilic archaeum Pyrococcus horikoshii OT3.
|
| |
Extremophiles,
9,
127-134.
|
 |
|
|
|
|
 |
D.Triantafillidou,
E.Persidou,
D.Lazarou,
P.Andrikopoulos,
F.Leontiadou,
and
T.Choli-Papadopoulou
(2004).
Structural destabilization of the recombinant thermophilic TthL11 ribosomal protein by a single amino acid substitution.
|
| |
Biol Chem,
385,
31-39.
|
 |
|
|
|
|
 |
I.Vakonakis,
J.Sun,
T.Wu,
A.Holzenburg,
S.S.Golden,
and
A.C.LiWang
(2004).
NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for KaiA-KaiC interaction.
|
| |
Proc Natl Acad Sci U S A,
101,
1479-1484.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Usui,
N.Ishii,
Y.Kawarabayasi,
and
M.Yohda
(2004).
Expression and biochemical characterization of two small heat shock proteins from the thermoacidophilic crenarchaeon Sulfolobus tokodaii strain 7.
|
| |
Protein Sci,
13,
134-144.
|
 |
|
|
|
|
 |
A.P.Lin,
and
L.McAlister-Henn
(2003).
Homologous binding sites in yeast isocitrate dehydrogenase for cofactor (NAD+) and allosteric activator (AMP).
|
| |
J Biol Chem,
278,
12864-12872.
|
 |
|
|
|
|
 |
J.Miyazaki,
N.Kobashi,
M.Nishiyama,
and
H.Yamane
(2003).
Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic bacterium, Thermus thermophilus HB27, and evolutionary implication of beta-decarboxylating dehydrogenase.
|
| |
J Biol Chem,
278,
1864-1871.
|
 |
|
|
|
|
 |
J.Sivaraman,
Y.Li,
J.Banks,
D.E.Cane,
A.Matte,
and
M.Cygler
(2003).
Crystal structure of Escherichia coli PdxA, an enzyme involved in the pyridoxal phosphate biosynthesis pathway.
|
| |
J Biol Chem,
278,
43682-43690.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Yasutake,
S.Watanabe,
M.Yao,
Y.Takada,
N.Fukunaga,
and
I.Tanaka
(2003).
Crystal structure of the monomeric isocitrate dehydrogenase in the presence of NADP+: insight into the cofactor recognition, catalysis, and evolution.
|
| |
J Biol Chem,
278,
36897-36904.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Inoue,
T.Tamura,
N.Ehara,
A.Nishito,
Y.Nakayama,
M.Maekawa,
K.Imada,
H.Tanaka,
and
K.Inagaki
(2002).
Biochemical and molecular characterization of the NAD(+)-dependent isocitrate dehydrogenase from the chemolithotroph Acidithiobacillus thiooxidans.
|
| |
FEMS Microbiol Lett,
214,
127-132.
|
 |
|
|
|
|
 |
M.Karlström,
I.H.Steen,
G.Tibbelin,
T.Lien,
N.K.Birkeland,
and
R.Ladenstein
(2002).
Crystallization and preliminary X-ray structure analysis of isocitrate dehydrogenase from two hyperthermophiles, Aeropyrum pernix and Thermotoga maritima.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
2162-2164.
|
 |
|
|
|
|
 |
C.Qu,
S.Akanuma,
N.Tanaka,
H.Moriyama,
and
T.Oshima
(2001).
Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
225-232.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.H.Steen,
D.Madern,
M.Karlström,
T.Lien,
R.Ladenstein,
and
N.K.Birkeland
(2001).
Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state, and phylogenetic affiliation.
|
| |
J Biol Chem,
276,
43924-43931.
|
 |
|
|
|
|
 |
K.A.Denessiouk,
V.V.Rantanen,
and
M.S.Johnson
(2001).
Adenine recognition: a motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins.
|
| |
Proteins,
44,
282-291.
|
 |
|
|
|
|
 |
K.Numata,
Y.Hayashi-Iwasaki,
J.Kawaguchi,
M.Sakurai,
H.Moriyama,
N.Tanaka,
and
T.Oshima
(2001).
Thermostabilization of a chimeric enzyme by residue substitutions: four amino acid residues in loop regions are responsible for the thermostability of Thermus thermophilus isopropylmalate dehydrogenase.
|
| |
Biochim Biophys Acta,
1545,
174-183.
|
 |
|
|
|
|
 |
M.Fujita,
H.Tamegai,
T.Eguchi,
and
K.Kakinuma
(2001).
Novel substrate specificity of designer 3-isopropylmalate dehydrogenase derived from Thermus thermophilus HB8.
|
| |
Biosci Biotechnol Biochem,
65,
2695-2700.
|
 |
|
|
|
|
 |
R.Chen,
and
S.S.Jeong
(2000).
Functional prediction: identification of protein orthologs and paralogs.
|
| |
Protein Sci,
9,
2344-2353.
|
 |
|
|
|
|
 |
S.A.Doyle,
S.Y.Fung,
and
D.E.Koshland
(2000).
Redesigning the substrate specificity of an enzyme: isocitrate dehydrogenase.
|
| |
Biochemistry,
39,
14348-14355.
|
 |
|
|
|
|
 |
C.Motono,
A.Yamagishi,
and
T.Oshima
(1999).
Urea-induced unfolding and conformational stability of 3-isopropylmalate dehydrogenase from the Thermophile thermus thermophilus and its mesophilic counterpart from Escherichia coli.
|
| |
Biochemistry,
38,
1332-1337.
|
 |
|
|
|
|
 |
R.Schleif
(1999).
Arm-domain interactions in proteins: a review.
|
| |
Proteins,
34,
1-3.
|
 |
|
|
|
|
 |
S.Akanuma,
A.Yamagishi,
N.Tanaka,
and
T.Oshima
(1999).
Further improvement of the thermal stability of a partially stabilized Bacillus subtilis 3-isopropylmalate dehydrogenase variant by random and site-directed mutagenesis.
|
| |
Eur J Biochem,
260,
499-504.
|
 |
|
|
|
|
 |
Y.Korkhin,
A.J.Kalb (Gilboa),
M.Peretz,
O.Bogin,
Y.Burstein,
and
F.Frolow
(1999).
Oligomeric integrity--the structural key to thermal stability in bacterial alcohol dehydrogenases.
|
| |
Protein Sci,
8,
1241-1249.
|
 |
|
|
|
|
 |
Y.Xu,
G.Bhargava,
H.Wu,
G.Loeber,
and
L.Tong
(1999).
Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: a new class of oxidative decarboxylases.
|
| |
Structure,
7,
R877-R889.
|
 |
|
|
|
|
 |
D.Tsuchiya,
and
A.Takenaka
(1998).
Romit profile analysis for molecular replacements.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
151-153.
|
 |
|
|
|
|
 |
H.Matsunami,
H.Kawaguchi,
K.Inagaki,
T.Eguchi,
K.Kakinuma,
and
H.Tanaka
(1998).
Overproduction and substrate specificity of 3-isopropylmalate dehydrogenase from Thiobacillus ferrooxidans.
|
| |
Biosci Biotechnol Biochem,
62,
372-373.
|
 |
|
|
|
|
 |
K.Imada,
K.Inagaki,
H.Matsunami,
H.Kawaguchi,
H.Tanaka,
N.Tanaka,
and
K.Namba
(1998).
Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
|
| |
Structure,
6,
971-982.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Závodszky,
J.Kardos,
Svingor,
and
G.A.Petsko
(1998).
Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins.
|
| |
Proc Natl Acad Sci U S A,
95,
7406-7411.
|
 |
|
|
|
|
 |
S.Akanuma,
A.Yamagishi,
N.Tanaka,
and
T.Oshima
(1998).
Serial increase in the thermal stability of 3-isopropylmalate dehydrogenase from Bacillus subtilis by experimental evolution.
|
| |
Protein Sci,
7,
698-705.
|
 |
|
|
|
|
 |
S.Kashiwabara,
Y.Matsuki,
T.Kishimoto,
and
Y.Suzuki
(1998).
Clustered proline residues around the active-site cleft in thermostable oligo-1,6-glucosidase of Bacillus flavocaldarius KP1228.
|
| |
Biosci Biotechnol Biochem,
62,
1093-1102.
|
 |
|
|
|
|
 |
T.Mikawa,
R.Kato,
M.Sugahara,
and
S.Kuramitsu
(1998).
Thermostable repair enzyme for oxidative DNA damage from extremely thermophilic bacterium, Thermus thermophilus HB8.
|
| |
Nucleic Acids Res,
26,
903-910.
|
 |
|
|
|
|
 |
T.Suzuki,
H.Moriyama,
R.Hirose,
M.Sakurai,
N.Tanaka,
and
T.Oshima
(1998).
Crystallization and preliminary X-ray studies on the hyperstable 3-isopropylmalate dehydrogenase from the thermoacidophilic archaeon Sulfolobus sp. strain 7.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
444-445.
|
 |
|
|
|
|
 |
A.M.Dean,
and
G.B.Golding
(1997).
Protein engineering reveals ancient adaptive replacements in isocitrate dehydrogenase.
|
| |
Proc Natl Acad Sci U S A,
94,
3104-3109.
|
 |
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
L.Prade,
P.Hof,
and
B.Bieseler
(1997).
Dimer interface of glutathione S-transferase from Arabidopsis thaliana: influence of the G-site architecture on the dimer interface and implications for classification.
|
| |
Biol Chem,
378,
317-320.
|
 |
|
|
|
|
 |
M.Hennig,
R.Sterner,
K.Kirschner,
and
J.N.Jansonius
(1997).
Crystal structure at 2.0 A resolution of phosphoribosyl anthranilate isomerase from the hyperthermophile Thermotoga maritima: possible determinants of protein stability.
|
| |
Biochemistry,
36,
6009-6016.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Van de Casteele,
P.Chen,
M.Roovers,
C.Legrain,
and
N.Glansdorff
(1997).
Structure and expression of a pyrimidine gene cluster from the extreme thermophile Thermus strain ZO5.
|
| |
J Bacteriol,
179,
3470-3481.
|
 |
|
|
|
|
 |
R.Chen,
A.F.Greer,
and
A.M.Dean
(1997).
Structural constraints in protein engineering--the coenzyme specificity of Escherichia coli isocitrate dehydrogenase.
|
| |
Eur J Biochem,
250,
578-582.
|
 |
|
|
|
|
 |
R.Kato,
K.Hasegawa,
Y.Hidaka,
S.Kuramitsu,
and
T.Hoshino
(1997).
Characterization of a thermostable DNA photolyase from an extremely thermophilic bacterium, Thermus thermophilus HB27.
|
| |
J Bacteriol,
179,
6499-6503.
|
 |
|
|
|
|
 |
T.Suzuki,
Y.Inoki,
A.Yamagishi,
T.Iwasaki,
T.Wakagi,
and
T.Oshima
(1997).
Molecular and phylogenetic characterization of isopropylmalate dehydrogenase of a thermoacidophilic archaeon, Sulfolobus sp. strain 7.
|
| |
J Bacteriol,
179,
1174-1179.
|
 |
|
|
|
|
 |
W.D.Kohn,
C.T.Mant,
and
R.S.Hodges
(1997).
Alpha-helical protein assembly motifs.
|
| |
J Biol Chem,
272,
2583-2586.
|
 |
|
|
|
|
 |
W.N.Zhao,
and
L.McAlister-Henn
(1997).
Affinity purification and kinetic analysis of mutant forms of yeast NAD+-specific isocitrate dehydrogenase.
|
| |
J Biol Chem,
272,
21811-21817.
|
 |
|
|
|
|
 |
J.H.Hurley,
R.Chen,
and
A.M.Dean
(1996).
Determinants of cofactor specificity in isocitrate dehydrogenase: structure of an engineered NADP+ --> NAD+ specificity-reversal mutant.
|
| |
Biochemistry,
35,
5670-5678.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Chen,
A.Greer,
and
A.M.Dean
(1996).
Redesigning secondary structure to invert coenzyme specificity in isopropylmalate dehydrogenase.
|
| |
Proc Natl Acad Sci U S A,
93,
12171-12176.
|
 |
|
|
|
|
 |
R.Chen,
J.A.Grobler,
J.H.Hurley,
and
A.M.Dean
(1996).
Second-site suppression of regulatory phosphorylation in Escherichia coli isocitrate dehydrogenase.
|
| |
Protein Sci,
5,
287-295.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Akanuma,
A.Yamagishi,
N.Tanaka,
and
T.Oshima
(1996).
Spontaneous tandem sequence duplications reverse the thermal stability of carboxyl-terminal modified 3-isopropylmalate dehydrogenase.
|
| |
J Bacteriol,
178,
6300-6304.
|
 |
|
|
|
|
 |
T.Kotsuka,
S.Akanuma,
M.Tomuro,
A.Yamagishi,
and
T.Oshima
(1996).
Further stabilization of 3-isopropylmalate dehydrogenase of an extreme thermophile, Thermus thermophilus, by a suppressor mutation method.
|
| |
J Bacteriol,
178,
723-727.
|
 |
|
|
|
|
 |
Y.Hayashi-Iwasaki,
K.Numata,
A.Yamagishi,
K.Yutani,
M.Sakurai,
N.Tanaka,
and
T.Oshima
(1996).
A stable intermediate in the thermal unfolding process of a chimeric 3-isopropylmalate dehydrogenase between a thermophilic and a mesophilic enzymes.
|
| |
Protein Sci,
5,
511-516.
|
 |
|
|
|
|
 |
Z.Y.Zhu,
and
S.Karlin
(1996).
Clusters of charged residues in protein three-dimensional structures.
|
| |
Proc Natl Acad Sci U S A,
93,
8350-8355.
|
 |
|
|
|
|
 |
A.M.Dean,
and
L.Dvorak
(1995).
The role of glutamate 87 in the kinetic mechanism of Thermus thermophilus isopropylmalate dehydrogenase.
|
| |
Protein Sci,
4,
2156-2167.
|
 |
|
|
|
|
 |
D.T.Logan,
M.H.Mazauric,
D.Kern,
and
D.Moras
(1995).
Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus.
|
| |
EMBO J,
14,
4156-4167.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Yoda,
Y.Anraku,
H.Kirino,
T.Wakagi,
and
T.Oshima
(1995).
Purification and characterization of 3-isopropylmalate dehydrogenase from a thermoacidophilic archaebacterium Sulfolobus sp. strain 7.
|
| |
FEMS Microbiol Lett,
131,
243-247.
|
 |
|
|
|
|
 |
M.Tamakoshi,
A.Yamagishi,
and
T.Oshima
(1995).
Screening of stable proteins in an extreme thermophile, Thermus thermophilus.
|
| |
Mol Microbiol,
16,
1031-1036.
|
 |
|
|
|
|
 |
P.Crouzet,
and
L.Otten
(1995).
Sequence and mutational analysis of a tartrate utilization operon from Agrobacterium vitis.
|
| |
J Bacteriol,
177,
6518-6526.
|
 |
|
|
|
|
 |
S.Kawaguchi,
and
S.Kuramitsu
(1995).
Separation of heat-stable proteins from Thermus thermophilus HB8 by two-dimensional electrophoresis.
|
| |
Electrophoresis,
16,
1060-1066.
|
 |
|
|
|
|
 |
T.Zhang,
and
D.E.Koshland
(1995).
Modeling substrate binding in Thermus thermophilus isopropylmalate dehydrogenase.
|
| |
Protein Sci,
4,
84-92.
|
 |
|
|
|
|
 |
H.Kirino,
M.Aoki,
M.Aoshima,
Y.Hayashi,
M.Ohba,
A.Yamagishi,
T.Wakagi,
and
T.Oshima
(1994).
Hydrophobic interaction at the subunit interface contributes to the thermostability of 3-isopropylmalate dehydrogenase from an extreme thermophile, Thermus thermophilus.
|
| |
Eur J Biochem,
220,
275-281.
|
 |
|
|
|
|
 |
J.H.Hurley,
and
A.M.Dean
(1994).
Structure of 3-isopropylmalate dehydrogenase in complex with NAD+: ligand-induced loop closing and mechanism for cofactor specificity.
|
| |
Structure,
2,
1007-1016.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Miyazaki,
T.Yaoi,
and
T.Oshima
(1994).
Expression, purification, and substrate specificity of isocitrate dehydrogenase from Thermus thermophilus HB8.
|
| |
Eur J Biochem,
221,
899-903.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

