 |
PDBsum entry 1imf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.1.3.25
- inositol-phosphate phosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a myo-inositol phosphate + H2O = myo-inositol + phosphate
|
 |
 |
 |
 |
 |
myo-inositol phosphate
|
+
|
H2O
|
=
|
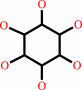 myo-inositol
myo-inositol
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.94
- D-galactose 1-phosphate phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-galactose 1-phosphate + H2O = D-galactose + phosphate
|
 |
 |
 |
 |
 |
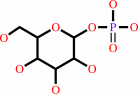 alpha-D-galactose 1-phosphate
alpha-D-galactose 1-phosphate
|
+
|
H2O
|
=
|
 D-galactose
D-galactose
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:9468-9476
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural studies of metal binding by inositol monophosphatase: evidence for two-metal ion catalysis.
|
|
R.Bone,
L.Frank,
J.P.Springer,
J.R.Atack.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of inositol monophosphatase has been determined to 2.60 A
resolution in complexes with Mn2+ and with Mn2+ and phosphate. In the Mn2+
complex, three metal cations and one Cl were bound in the active site on each of
the two subunits of the enzyme. Ligands to the three metals include the side
chains of Glu 70, Asp 90, Asp 93, and Asp 220, t he carbonyl group of Ile 92,
several solvent molecules and the chloride, which is a ligand to each of the
cations. When phosphate is soaked into these Mn2+ cocrystals, one of the three
Mn2+ ions is expelled from the active site, leaving metal ions with octahedral
and tetrahedral coordination geometry. In addition, the structure of apoinositol
monophosphatase was determined to 2.5 A resolution. Residues 70-75, a two-turn
helical segment which is involved in metal coordination, moves away from the
metal binding site by 2-3 A in the absence of cations. Residues 30-40, which
wrap around the metal binding site and interact with the metal indirectly
through solvent molecules and protein ligands to the metal, become disordered in
the absence of metal. In various metal complexes, segmental mobility is also
observed in the residues which form the metal binding sites. The results of
these studies of the interaction of inositol monophosphatase with cations
suggest that the enzyme accomplishes phosphate ester hydrolysis using two metal
ions, one with octahedral and one with tetrahedral coordination geometry. Broad
metal-binding specificity appears to result from extensive flexibility in
several of the protein segments which contribute metal ligands, from the
presence of alternate metal ligands and from metal coordination spheres which
include water molecules.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Pasquali,
C.L.Busceti,
F.Fulceri,
A.Paparelli,
and
F.Fornai
(2010).
Intracellular pathways underlying the effects of lithium.
|
| |
Behav Pharmacol,
21,
473-492.
|
 |
|
|
|
|
 |
Y.G.Lee,
S.G.Kang,
J.H.Lee,
S.I.Kim,
and
Y.H.Chung
(2010).
Characterization of hyperthermostable fructose-1,6-bisphosphatase from Thermococcus onnurineus NA1.
|
| |
J Microbiol,
48,
803-807.
|
 |
|
|
|
|
 |
Z.Li,
K.A.Stieglitz,
A.L.Shrout,
Y.Wei,
R.M.Weis,
B.Stec,
and
M.F.Roberts
(2010).
Mobile loop mutations in an archaeal inositol monophosphatase: modulating three-metal ion assisted catalysis and lithium inhibition.
|
| |
Protein Sci,
19,
309-318.
|
 |
|
|
|
|
 |
G.Brown,
A.Singer,
V.V.Lunin,
M.Proudfoot,
T.Skarina,
R.Flick,
S.Kochinyan,
R.Sanishvili,
A.Joachimiak,
A.M.Edwards,
A.Savchenko,
and
A.F.Yakunin
(2009).
Structural and biochemical characterization of the type II fructose-1,6-bisphosphatase GlpX from Escherichia coli.
|
| |
J Biol Chem,
284,
3784-3792.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.K.Brown,
G.Meng,
H.Ghadbane,
D.J.Scott,
L.G.Dover,
J.Nigou,
G.S.Besra,
and
K.Fütterer
(2007).
Dimerization of inositol monophosphatase Mycobacterium tuberculosis SuhB is not constitutive, but induced by binding of the activator Mg2+.
|
| |
BMC Struct Biol,
7,
55.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Arai,
K.Ito,
T.Ohnishi,
H.Ohba,
R.Akasaka,
Y.Bessho,
K.Hanawa-Suetsugu,
T.Yoshikawa,
M.Shirouzu,
and
S.Yokoyama
(2007).
Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures.
|
| |
Proteins,
67,
732-742.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Gill,
F.Mohammed,
R.Badyal,
L.Coates,
P.Erskine,
D.Thompson,
J.Cooper,
M.Gore,
and
S.Wood
(2005).
High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
545-555.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Nishimasu,
S.Fushinobu,
H.Shoun,
and
T.Wakagi
(2004).
The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
|
| |
Structure,
12,
949-959.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.A.Stieglitz,
K.A.Johnson,
H.Yang,
M.F.Roberts,
B.A.Seaton,
J.F.Head,
and
B.Stec
(2002).
Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
|
| |
J Biol Chem,
277,
22863-22874.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.J.Phiel,
and
P.S.Klein
(2001).
Molecular targets of lithium action.
|
| |
Annu Rev Pharmacol Toxicol,
41,
789-813.
|
 |
|
|
|
|
 |
J.W.Pettegrew,
K.Panchalingam,
R.J.McClure,
S.Gershon,
L.R.Muenz,
and
J.Levine
(2001).
Effects of chronic lithium administration on rat brain phosphatidylinositol cycle constituents, membrane phospholipids and amino acids.
|
| |
Bipolar Disord,
3,
189-201.
|
 |
|
|
|
|
 |
K.A.Johnson,
L.Chen,
H.Yang,
M.F.Roberts,
and
B.Stec
(2001).
Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
|
| |
Biochemistry,
40,
618-630.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.J.Miller,
M.W.Beaton,
J.Wilkie,
and
D.Gani
(2000).
The 6-OH group of D-inositol 1-phosphate serves as an H-bond donor in the catalytic hydrolysis of the phosphate ester by inositol monophosphatase.
|
| |
Chembiochem,
1,
262-271.
|
 |
|
|
|
|
 |
L.Chen,
and
M.F.Roberts
(2000).
Overexpression, purification, and analysis of complementation behavior of E. coli SuhB protein: comparison with bacterial and archaeal inositol monophosphatases.
|
| |
Biochemistry,
39,
4145-4153.
|
 |
|
|
|
|
 |
X.Zhou,
F.Alber,
G.Folkers,
G.H.Gonnet,
and
G.Chelvanayagam
(2000).
An analysis of the helix-to-strand transition between peptides with identical sequence.
|
| |
Proteins,
41,
248-256.
|
 |
|
|
|
|
 |
B.D.Spiegelberg,
J.P.Xiong,
J.J.Smith,
R.F.Gu,
and
J.D.York
(1999).
Cloning and characterization of a mammalian lithium-sensitive bisphosphate 3'-nucleotidase inhibited by inositol 1,4-bisphosphate.
|
| |
J Biol Chem,
274,
13619-13628.
|
 |
|
|
|
|
 |
D.E.Timm,
H.A.Mueller,
P.Bhanumoorthy,
J.M.Harp,
and
G.J.Bunick
(1999).
Crystal structure and mechanism of a carbon-carbon bond hydrolase.
|
| |
Structure,
7,
1023-1033.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Chen,
and
M.F.Roberts
(1999).
Characterization of a tetrameric inositol monophosphatase from the hyperthermophilic bacterium Thermotoga maritima.
|
| |
Appl Environ Microbiol,
65,
4559-4567.
|
 |
|
|
|
|
 |
S.Shan,
A.Yoshida,
S.Sun,
J.A.Piccirilli,
and
D.Herschlag
(1999).
Three metal ions at the active site of the Tetrahymena group I ribozyme.
|
| |
Proc Natl Acad Sci U S A,
96,
12299-12304.
|
 |
|
|
|
|
 |
L.Chen,
and
M.F.Roberts
(1998).
Cloning and expression of the inositol monophosphatase gene from Methanococcus jannaschii and characterization of the enzyme.
|
| |
Appl Environ Microbiol,
64,
2609-2615.
|
 |
|
|
|
|
 |
M.V.Ellis,
S.R.James,
O.Perisic,
C.P.Downes,
R.L.Williams,
and
M.Katan
(1998).
Catalytic domain of phosphoinositide-specific phospholipase C (PLC). Mutational analysis of residues within the active site and hydrophobic ridge of plcdelta1.
|
| |
J Biol Chem,
273,
11650-11659.
|
 |
|
|
|
|
 |
J.R.Atack
(1997).
Inositol monophosphatase inhibitors--lithium mimetics?
|
| |
Med Res Rev,
17,
215-224.
|
 |
|
|
|
|
 |
K.Rees-Milton,
M.Thorne,
P.Greasley,
J.Churchich,
and
M.G.Gore
(1997).
Detection of metal binding to bovine inositol monophosphatase by changes in the near and far ultraviolet regions of the CD spectrum.
|
| |
Eur J Biochem,
246,
211-217.
|
 |
|
|
|
|
 |
A.J.Ganzhorn,
P.Lepage,
P.D.Pelton,
F.Strasser,
P.Vincendon,
and
J.M.Rondeau
(1996).
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase.
|
| |
Biochemistry,
35,
10957-10966.
|
 |
|
|
|
|
 |
F.Moreno,
S.Corrales,
F.Garcia Blanco,
M.G.Gore,
K.Rees-Milton,
and
J.E.Churchich
(1996).
Reversible denaturation of myo-inositol monophosphatase. The stability of the metal-binding loop.
|
| |
Eur J Biochem,
240,
435-442.
|
 |
|
|
|
|
 |
V.Saudek,
P.Vincendon,
Q.T.Do,
R.A.Atkinson,
V.Sklenar,
P.D.Pelton,
F.Piriou,
and
A.J.Ganzhorn
(1996).
7Li nuclear-magnetic-resonance study of lithium binding to myo-inositolmonophosphatase.
|
| |
Eur J Biochem,
240,
288-291.
|
 |
|
|
|
|
 |
J.R.Atack
(1995).
Inositol monophosphatase inhibitors: a novel treatment for bipolar disorder?
|
| |
Biol Psychiatry,
37,
761-763.
|
 |
|
|
|
|
 |
V.Villeret,
S.Huang,
H.J.Fromm,
and
W.N.Lipscomb
(1995).
Crystallographic evidence for the action of potassium, thallium, and lithium ions on fructose-1,6-bisphosphatase.
|
| |
Proc Natl Acad Sci U S A,
92,
8916-8920.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

