 |
PDBsum entry 1iji

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.9
- histidinol-phosphate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Histidine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-histidinol phosphate + 2-oxoglutarate = 3-(imidazol-4-yl)-2-oxopropyl phosphate + L-glutamate
|
 |
 |
 |
 |
 |
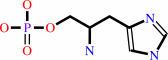 L-histidinol phosphate
L-histidinol phosphate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
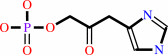 3-(imidazol-4-yl)-2-oxopropyl phosphate
3-(imidazol-4-yl)-2-oxopropyl phosphate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
311:761-776
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
|
|
J.Sivaraman,
Y.Li,
R.Larocque,
J.D.Schrag,
M.Cygler,
A.Matte.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The biosynthesis of histidine is a central metabolic process in organisms
ranging from bacteria to yeast and plants. The seventh step in the synthesis of
histidine within eubacteria is carried out by a pyridoxal-5'-phosphate
(PLP)-dependent l-histidinol phosphate aminotransferase (HisC, EC 2.6.1.9).
Here, we report the crystal structure of l-histidinol phosphate aminotransferase
from Escherichia coli, as a complex with pyridoxamine-5'-phosphate (PMP) at 1.5
A resolution, as the internal aldimine with PLP, and in a covalent, tetrahedral
complex consisting of PLP and l-histidinol phosphate attached to Lys214, both at
2.2 A resolution. This covalent complex resembles, in structural terms, the
gem-diamine intermediate that is formed transiently during conversion of the
internal to external aldimine.HisC is a dimeric enzyme with a mass of
approximately 80 kDa. Like most PLP-dependent enzymes, each HisC monomer
consists of two domains, a larger PLP-binding domain having an alpha/beta/alpha
topology, and a smaller domain. An N-terminal arm contributes to the
dimerization of the two monomers. The PLP-binding domain of HisC shows weak
sequence similarity, but significant structural similarity with the PLP-binding
domains of a number of PLP-dependent enzymes. Residues that interact with the
PLP cofactor, including Tyr55, Asn157, Asp184, Tyr187, Ser213, Lys214 and
Arg222, are conserved in the family of aspartate, tyrosine and histidinol
phosphate aminotransferases. The imidazole ring of l-histidinol phosphate is
bound, in part, through a hydrogen bond with Tyr110, a residue that is
substituted by Phe in the broad substrate specific HisC enzymes from Zymomonas
mobilis and Bacillus subtilis.Comparison of the structures of the HisC internal
aldimine, the PMP complex and the HisC l-histidinol phosphate complex reveal
minimal changes in protein or ligand structure. Proton transfer, required for
conversion of the gem-diamine to the external aldimine, does not appear to be
limited by the distance between substrate and lysine amino groups. We propose
that the tetrahedral complex has resulted from non-productive binding of
l-histidinol phosphate soaked into the HisC crystals, resulting in its inability
to be converted to the external aldimine at the HisC active site.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 7.
Figure 7. Schematic views of HisC-ligand interactions for
(a) the complex of HisC with PMP (b) the HisC internal aldimine
with PLP (Llp) and (c) the covalent complex of HisC with PLP and
Image -histidinol phosphate (Lph). Hydrogen bonds between HisC
and the ligands (less than or equal to 3.2 Å) are
indicated by broken lines. These Figures were prepared using the
program LIGPLOT.[56]
|
 |
Figure 9.
Figure 9. Schematic diagram showing the structures from the
transimination portion of the aminotransferase reaction
mechanism. The structures depicted are for (I) internal
aldimine, (II) gem-diamine intermediate protonated at substrate
N, (III) gem-diamine intermediate protonated at lysine NZ, and
(IV) external aldimine.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2001,
311,
761-776)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Marienhagen,
T.Sandalova,
H.Sahm,
L.Eggeling,
and
G.Schneider
(2008).
Insights into the structural basis of substrate recognition by histidinol-phosphate aminotransferase from Corynebacterium glutamicum.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
675-685.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Lima,
R.Khristoforov,
C.Momany,
and
R.S.Phillips
(2007).
Crystal structure of Homo sapiens kynureninase.
|
| |
Biochemistry,
46,
2735-2744.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Paiardini,
F.Bossa,
and
S.Pascarella
(2004).
Evolutionarily conserved regions and hydrophobic contacts at the superfamily level: The case of the fold-type I, pyridoxal-5'-phosphate-dependent enzymes.
|
| |
Protein Sci,
13,
2992-3005.
|
 |
|
|
|
|
 |
A.Matte,
J.Sivaraman,
I.Ekiel,
K.Gehring,
Z.Jia,
and
M.Cygler
(2003).
Contribution of structural genomics to understanding the biology of Escherichia coli.
|
| |
J Bacteriol,
185,
3994-4002.
|
 |
|
|
|
|
 |
E.S.Rangarajan,
J.Sivaraman,
A.Matte,
and
M.Cygler
(2002).
Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli.
|
| |
Proteins,
48,
737-740.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Barbosa,
J.Sivaraman,
Y.Li,
R.Larocque,
A.Matte,
J.D.Schrag,
and
M.Cygler
(2002).
Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
|
| |
Proc Natl Acad Sci U S A,
99,
1859-1864.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

