 |
PDBsum entry 1i2r

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.2.1.2
- transaldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-erythrose 4-phosphate + beta-D-fructose 6-phosphate
|
 |
 |
 |
 |
 |
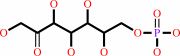 D-sedoheptulose 7-phosphate
D-sedoheptulose 7-phosphate
|
+
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
=
|
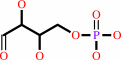 D-erythrose 4-phosphate
D-erythrose 4-phosphate
|
+
|
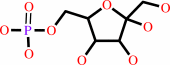 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Eur J Biochem
268:2408-2415
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Identification of catalytically important residues in the active site of Escherichia coli transaldolase.
|
|
U.Schörken,
S.Thorell,
M.Schürmann,
J.Jia,
G.A.Sprenger,
G.Schneider.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The roles of invariant residues at the active site of transaldolase B from
Escherichia coli have been probed by site-directed mutagenesis. The mutant
enzymes D17A, N35A, E96A, T156A, and S176A were purified from a talB-deficient
host and analyzed with respect to their 3D structure and kinetic behavior. X-ray
analysis showed that side chain replacement did not induce unanticipated
structural changes in the mutant enzymes. Three mutations, N35A, E96A, and T156A
resulted mainly in an effect on apparent kcat, with little changes in apparent
Km values for the substrates. Residues N35 and T156 are involved in the
positioning of a catalytic water molecule at the active site and the side chain
of E96 participates in concert with this water molecule in proton transfer
during catalysis. Substitution of Ser176 by alanine resulted in a mutant enzyme
with 2.5% residual activity. The apparent Km value for the donor substrate,
fructose 6-phosphate, was increased nearly fivefold while the apparent Km value
for the acceptor substrate, erythrose 4-phosphate remained unchanged, consistent
with a function for S176 in the binding of the C1 hydroxyl group of the donor
substrate. The mutant D17A showed a 300-fold decrease in kcat, and a fivefold
increase in the apparent Km value for the acceptor substrate erythrose
4-phosphate, suggesting a role of this residue in carbon-carbon bond cleavage
and stabilization of the carbanion/enamine intermediate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Stereo views of the final 2|Fo|-|Fc| electron
density maps, contoured at 1  , of the
transaldolase mutants D17A (A) and S176A (B). , of the
transaldolase mutants D17A (A) and S176A (B).
|
 |
Figure 4.
Fig. 4. Proposed reaction mechanism of transaldolase. The
steps leading to the central carbanion/enamine intermediate are
shown. The second half of the reaction, the addition of the
acceptor substrate is in principle the reverse of the first half
of the catalytic cycle and is therefore not included in the
figure. For sake of clarity, only conserved amino-acid side
chains proposed to participate in proton transfer during the
reaction are shown.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Federation of European Biochemical Societies:
Eur J Biochem
(2001,
268,
2408-2415)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Samland,
and
G.A.Sprenger
(2009).
Transaldolase: from biochemistry to human disease.
|
| |
Int J Biochem Cell Biol,
41,
1482-1494.
|
 |
|
|
|
|
 |
H.Huang,
H.Rong,
X.Li,
S.Tong,
Z.Zhu,
L.Niu,
and
M.Teng
(2008).
The crystal structure and identification of NQM1/YGR043C, a transaldolase from Saccharomyces cerevisiae.
|
| |
Proteins,
73,
1076-1081.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Schneider,
T.Sandalova,
G.Schneider,
G.A.Sprenger,
and
A.K.Samland
(2008).
Replacement of a Phenylalanine by a Tyrosine in the Active Site Confers Fructose-6-phosphate Aldolase Activity to the Transaldolase of Escherichia coli and Human Origin.
|
| |
J Biol Chem,
283,
30064-30072.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.La,
and
D.R.Livesay
(2005).
Predicting functional sites with an automated algorithm suitable for heterogeneous datasets.
|
| |
BMC Bioinformatics,
6,
116.
|
 |
|
|
|
|
 |
M.Caillau,
and
W.Paul Quick
(2005).
New insights into plant transaldolase.
|
| |
Plant J,
43,
1.
|
 |
|
|
|
|
 |
M.St-Jean,
J.Lafrance-Vanasse,
B.Liotard,
and
J.Sygusch
(2005).
High resolution reaction intermediates of rabbit muscle fructose-1,6-bisphosphate aldolase: substrate cleavage and induced fit.
|
| |
J Biol Chem,
280,
27262-27270.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.J.Kleijn,
W.A.van Winden,
W.M.van Gulik,
and
J.J.Heijnen
(2005).
Revisiting the 13C-label distribution of the non-oxidative branch of the pentose phosphate pathway based upon kinetic and genetic evidence.
|
| |
FEBS J,
272,
4970-4982.
|
 |
|
|
|
|
 |
T.Soderberg,
and
R.C.Alver
(2004).
Transaldolase of Methanocaldococcus jannaschii.
|
| |
Archaea,
1,
255-262.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

