 |
PDBsum entry 1hqo

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Signaling protein
|
PDB id
|
|

|

|
1hqo
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.11.1.9
- glutathione peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 glutathione + H2O2 = glutathione disulfide + 2 H2O
|
 |
 |
 |
 |
 |
 2
×
glutathione
2
×
glutathione
|
+
|
 H2O2
H2O2
|
=
|
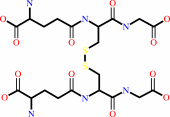 glutathione disulfide
glutathione disulfide
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Se(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.8.4.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
98:1459-1464
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of the nitrogen regulation fragment of the yeast prion protein Ure2p.
|
|
T.C.Umland,
K.L.Taylor,
S.Rhee,
R.B.Wickner,
D.R.Davies.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
is due to a prion form of the nitrogen
regulatory protein Ure2p. It is a negative regulator of nitrogen catabolism and
acts by inhibiting the transcription factor Gln3p. Ure2p residues 1--80 are
necessary for prion generation and propagation. The C-terminal fragment retains
nitrogen regulatory activity, albeit somewhat less efficiently than the
full-length protein, and it also lowers the frequency of prion generation. The
crystal structure of this C-terminal fragment, Ure2p(97--354), at 2.3 A
resolution is described here. It adopts the same fold as the glutathione
S-transferase superfamily, consistent with their sequence similarity. However,
Ure2p(97--354) lacks a properly positioned catalytic residue that is required
for S-transferase activity. Residues within this regulatory fragment that have
been indicated by mutational studies to influence prion generation have been
mapped onto the three-dimensional structure, and possible implications for prion
activity are discussed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Stereoview of the Ure2p(97-354). Monomer A is
green and Monomer B is cyan. Prion-inhibiting regions (His-151
to Ser-158 and Val-347 to Glu-354) are indicated in blue, and
the prion-promoting region (Ser-221 to Ile-227) is indicated in
red. Gold labels the position of two additional residues
implicated in affecting prion-induction, K127 and V271.
|
 |
Figure 4.
Fig. 4. The superposition of Monomer A of Ure2p(97-354)
(red) and a monomer of A. thaliana GST (38) (blue). This
representation is viewed into the cleft between the two domains,
which in GST contains the G- and H-sites.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Chen,
L.J.Chen,
H.Y.Wang,
Y.Q.Wang,
and
S.Perrett
(2011).
Deletion of a Ure2 C-terminal prion-inhibiting region promotes the rate of fibril seed formation and alters interaction with Hsp40.
|
| |
Protein Eng Des Sel,
24,
69-78.
|
 |
|
|
|
|
 |
U.Baxa,
P.W.Keller,
N.Cheng,
J.S.Wall,
and
A.C.Steven
(2011).
In Sup35p filaments (the [PSI+] prion), the globular C-terminal domains are widely offset from the amyloid fibril backbone.
|
| |
Mol Microbiol,
79,
523-532.
|
 |
|
|
|
|
 |
Y.Q.Wang,
M.Bongiovanni,
S.L.Gras,
and
S.Perrett
(2011).
The fibrils of Ure2p homologs from Saccharomyces cerevisiae and Saccharoymyces paradoxus have similar cross-β structure in both dried and hydrated forms.
|
| |
J Struct Biol,
174,
505-511.
|
 |
|
|
|
|
 |
Y.Yu,
H.Y.Wang,
M.Bai,
and
S.Perrett
(2011).
Flexibility of the Ure2 prion domain is important for amyloid fibril formation.
|
| |
Biochem J,
434,
143-151.
|
 |
|
|
|
|
 |
A.Böckmann,
and
B.H.Meier
(2010).
Prions: En route from structural models to structures.
|
| |
Prion,
4,
72-79.
|
 |
|
|
|
|
 |
C.Zhang,
A.P.Jackson,
Z.R.Zhang,
Y.Han,
S.Yu,
R.Q.He,
and
S.Perrett
(2010).
Amyloid-like aggregates of the yeast prion protein ure2 enter vertebrate cells by specific endocytotic pathways and induce apoptosis.
|
| |
PLoS One,
5,
0.
|
 |
|
|
|
|
 |
L.Fei,
and
S.Perrett
(2009).
Disulfide bond formation significantly accelerates the assembly of Ure2p fibrils because of the proximity of a potential amyloid stretch.
|
| |
J Biol Chem,
284,
11134-11141.
|
 |
|
|
|
|
 |
L.Pieri,
M.Bucciantini,
P.Guasti,
J.Savistchenko,
R.Melki,
and
M.Stefani
(2009).
Synthetic lipid vesicles recruit native-like aggregates and affect the aggregation process of the prion Ure2p: insights on vesicle permeabilization and charge selectivity.
|
| |
Biophys J,
96,
3319-3330.
|
 |
|
|
|
|
 |
C.Gancedo,
and
C.L.Flores
(2008).
Moonlighting proteins in yeasts.
|
| |
Microbiol Mol Biol Rev,
72,
197.
|
 |
|
|
|
|
 |
K.H.Wong,
M.J.Hynes,
and
M.A.Davis
(2008).
Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi.
|
| |
Eukaryot Cell,
7,
917-925.
|
 |
|
|
|
|
 |
F.Shewmaker,
L.Mull,
T.Nakayashiki,
D.C.Masison,
and
R.B.Wickner
(2007).
Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae.
|
| |
Genetics,
176,
1557-1565.
|
 |
|
|
|
|
 |
B.Ono,
M.Kubota,
H.Kimiduka,
H.Kawaminami,
T.Ueto,
S.Yokosawa,
M.Iseda,
Y.Yamamoto,
Y.Murakami,
and
S.Yokota
(2006).
Production of a polymer-forming fusion protein in Escerichia coli strain BL21.
|
| |
Biosci Biotechnol Biochem,
70,
2813-2823.
|
 |
|
|
|
|
 |
N.Ranson,
T.Stromer,
L.Bousset,
R.Melki,
and
L.C.Serpell
(2006).
Insights into the architecture of the Ure2p yeast protein assemblies from helical twisted fibrils.
|
| |
Protein Sci,
15,
2481-2487.
|
 |
|
|
|
|
 |
D.Davies,
and
D.Davies
(2005).
A quiet life with proteins.
|
| |
Annu Rev Biophys Biomol Struct,
34,
1.
|
 |
|
|
|
|
 |
J.C.Chan,
N.A.Oyler,
W.M.Yau,
and
R.Tycko
(2005).
Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10-39 of the yeast prion protein Ure2p.
|
| |
Biochemistry,
44,
10669-10680.
|
 |
|
|
|
|
 |
N.Talarek,
L.Maillet,
C.Cullin,
and
M.Aigle
(2005).
The [URE3] prion is not conserved among Saccharomyces species.
|
| |
Genetics,
171,
23-34.
|
 |
|
|
|
|
 |
R.Rai,
and
T.G.Cooper
(2005).
In vivo specificity of Ure2 protection from heavy metal ion and oxidative cellular damage in Saccharomyces cerevisiae.
|
| |
Yeast,
22,
343-358.
|
 |
|
|
|
|
 |
S.Catharino,
J.Buchner,
and
S.Walter
(2005).
Characterization of oligomeric species in the fibrillization pathway of the yeast prion Ure2p.
|
| |
Biol Chem,
386,
633-641.
|
 |
|
|
|
|
 |
A.V.Kajava,
U.Baxa,
R.B.Wickner,
and
A.C.Steven
(2004).
A model for Ure2p prion filaments and other amyloids: the parallel superpleated beta-structure.
|
| |
Proc Natl Acad Sci U S A,
101,
7885-7890.
|
 |
|
|
|
|
 |
E.D.Ross,
U.Baxa,
and
R.B.Wickner
(2004).
Scrambled prion domains form prions and amyloid.
|
| |
Mol Cell Biol,
24,
7206-7213.
|
 |
|
|
|
|
 |
R.B.Wickner,
H.K.Edskes,
E.D.Ross,
M.M.Pierce,
F.Shewmaker,
U.Baxa,
and
A.Brachmann
(2004).
Prions of yeast are genes made of protein: amyloids and enzymes.
|
| |
Cold Spring Harb Symp Quant Biol,
69,
489-496.
|
 |
|
|
|
|
 |
R.B.Wickner,
H.K.Edskes,
E.D.Ross,
M.M.Pierce,
U.Baxa,
A.Brachmann,
and
F.Shewmaker
(2004).
Prion genetics: new rules for a new kind of gene.
|
| |
Annu Rev Genet,
38,
681-707.
|
 |
|
|
|
|
 |
A.Baudin-Baillieu,
E.Fernandez-Bellot,
F.Reine,
E.Coissac,
and
C.Cullin
(2003).
Conservation of the prion properties of Ure2p through evolution.
|
| |
Mol Biol Cell,
14,
3449-3458.
|
 |
|
|
|
|
 |
H.K.Edskes,
and
R.B.Wickner
(2002).
Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full-length protein.
|
| |
Proc Natl Acad Sci U S A,
99,
16384-16391.
|
 |
|
|
|
|
 |
L.Bousset,
N.H.Thomson,
S.E.Radford,
and
R.Melki
(2002).
The yeast prion Ure2p retains its native alpha-helical conformation upon assembly into protein fibrils in vitro.
|
| |
EMBO J,
21,
2903-2911.
|
 |
|
|
|
|
 |
S.M.Uptain,
and
S.Lindquist
(2002).
Prions as protein-based genetic elements.
|
| |
Annu Rev Microbiol,
56,
703-741.
|
 |
|
|
|
|
 |
U.Baxa,
V.Speransky,
A.C.Steven,
and
R.B.Wickner
(2002).
Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein.
|
| |
Proc Natl Acad Sci U S A,
99,
5253-5260.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

