 |
PDBsum entry 1hoo

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.4
- adenylosuccinate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + L-aspartate + GTP = N6-(1,2-dicarboxyethyl)-AMP + GDP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
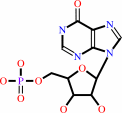 IMP
IMP
|
+
|
 L-aspartate
L-aspartate
|
+
|
 GTP
GTP
|
=
|
N(6)-(1,2-dicarboxyethyl)-AMP
Bound ligand (Het Group name = )
matches with 93.10% similarity
|
+
|
 GDP
GDP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
271:15407-15413
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Refined crystal structures of guanine nucleotide complexes of adenylosuccinate synthetase from Escherichia coli.
|
|
B.W.Poland,
Z.Hou,
C.Bruns,
H.J.Fromm,
R.B.Honzatko.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Structures of adenylosuccinate synthetase from Escherichia coli complexed with
guanosine-5'-(beta,gamma-imido) triphosphate and
guanosine-5'-(beta,gamma-methylene)triphosphate in the presence and the absence
of Mg2+ have been refined to R-factors below 0.2 against data to a nominal
resolution of 2.7 A. Asp333 of the synthetase hydrogen bonds to the exocyclic
2-amino and endocyclic N1 groups of the guanine nucleotide base, whereas the
hydroxyl of Ser414 and the backbone amide of Lys331 hydrogen bond to the 6-oxo
position. The side chains of Lys331 and Pro417 pack against opposite faces of
the guanine nucleotide base. The synthetase recognizes neither the N7 position
of guanine nucleotides nor the ribose group. Electron density for the
guanine-5'-(beta,gamma-imido) triphosphate complex is consistent with a mixture
of the triphosphate nucleoside and its hydrolyzed diphosphate nucleoside bound
to the active site. The base, ribose, and alpha-phosphate positions overlap, but
the beta-phosphates occupy different binding sites. The binding of
guanosine-5'-(beta,gamma-methylene)triphosphate to the active site is comparable
with that of guanosine-5'-(beta, gamma-imido)triphosphate. No electron density,
however, for the corresponding diphosphate nucleoside is observed. In addition,
electron density for bound Mg2+ is absent in these nucleotide complexes. The
guanine nucleotide complexes of the synthetase are compared with complexes of
other GTP-binding proteins and to a preliminary structure of the complex of GDP,
IMP, Mg2+, and succinate with the synthetase. The enzyme, under conditions
reported here, does not undergo a conformational change in response to the
binding of guanine nucleotides, and minimally IMP and/or Mg2+ must be present in
order to facilitate the complete recognition of the guanine nucleotide by the
synthetase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Stereoview of GppCp (bold lines) at its site of
ligation to adenylosuccinate synthetase. Top, an overview
representing the protein as a trace of its  -carbons.
Bottom, a detailed view of the region of binding of the guanine
nucleotide. -carbons.
Bottom, a detailed view of the region of binding of the guanine
nucleotide.
|
 |
Figure 3.
Fig. 3. Superposition of GppN, GppNp, and GppCp in the
conformations observed for these nucleotides in their ligand
complexes with the synthetase.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(1996,
271,
15407-15413)
copyright 1996.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.Lee,
A.Gorrell,
H.J.Fromm,
and
R.F.Colman
(1999).
Implication of arginine-131 and arginine-303 in the substrate site of adenylosuccinate synthetase of Escherichia coli by affinity labeling with 6-(4-bromo-2,3-dioxobutyl)thioadenosine 5'-monophosphate.
|
| |
Biochemistry,
38,
5754-5763.
|
 |
|
|
|
|
 |
B.W.Poland,
S.F.Lee,
M.V.Subramanian,
D.L.Siehl,
R.J.Anderson,
H.J.Fromm,
and
R.B.Honzatko
(1996).
Refined crystal structure of adenylosuccinate synthetase from Escherichia coli complexed with hydantocidin 5'-phosphate, GDP, HPO4(2-), Mg2+, and hadacidin.
|
| |
Biochemistry,
35,
15753-15759.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

