 |
PDBsum entry 1hfb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of the tyrosine-regulated 3-deoxy-d-arabino- heptulosonate-7-phosphate synthase from saccharomyces cerevisiae complexed with phosphoenolpyruvate
|
|
Structure:
|
 |
Tyrosine-regulated 3-deoxy-d-arabino-heptulosonate-7- phosphate synthase. Chain: a, b, c, d, e, f, g, h. Synonym: phospho-2-keto-3-deoxyheptonate aldolase, dahp synthetase, 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase, phospho-2- dehydro-3-deoxyheptonate aldolase tyrosine-inhibited, phospho-2- dehydro-3-deoxyheptonate aldolase. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Organism_taxid: 4932. Strain: rh1326. Expressed in: saccharomyces cerevisiae. Expression_system_taxid: 4932.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.208
|
R-free:
|
0.261
|
|
|
Authors:
|
 |
T.R.Schneider,M.Hartmann,G.H.Braus
|
Key ref:

|
 |
M.Hartmann
et al.
(2003).
Evolution of feedback-inhibited beta /alpha barrel isoenzymes by gene duplication and a single mutation.
Proc Natl Acad Sci U S A,
100,
862-867.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
30-Nov-00
|
Release date:
|
14-Jan-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P32449
(AROG_YEAST) -
Phospho-2-dehydro-3-deoxyheptonate aldolase, tyrosine-inhibited from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
370 a.a.
339 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.54
- 3-deoxy-7-phosphoheptulonate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Shikimate and Chorismate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-erythrose 4-phosphate + phosphoenolpyruvate + H2O = 7-phospho-2- dehydro-3-deoxy-D-arabino-heptonate + phosphate
|
 |
 |
 |
 |
 |
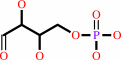 D-erythrose 4-phosphate
D-erythrose 4-phosphate
|
+
|
 phosphoenolpyruvate
phosphoenolpyruvate
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
7-phospho-2- dehydro-3-deoxy-D-arabino-heptonate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
100:862-867
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Evolution of feedback-inhibited beta /alpha barrel isoenzymes by gene duplication and a single mutation.
|
|
M.Hartmann,
T.R.Schneider,
A.Pfeil,
G.Heinrich,
W.N.Lipscomb,
G.H.Braus.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The betaalpha barrel is the common protein fold of numerous enzymes and was
proposed recently to be the result of gene duplication and fusion of an ancient
half-barrel. The initial enzyme of shikimate biosynthesis possesses the
additional feature of feedback regulation. The crystal structure and kinetic
studies on chimera and mutant proteins of yeast
3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP) synthase from Saccharomyces
cerevisiae inhibited by phenylalanine (Aro3p) and DAHP synthase S. cerevisiae
inhibited by tyrosine (Aro4p) give insight into important regions for regulation
in the enzyme: The loop, which is connecting the two half-barrels, and
structural elements added to the barrel are prerequisites for regulation and
form a cavity on the N-terminal side of the betaalpha barrel. In the cavity of
Aro4p at position 226 is a glycine residue, which is highly conserved in all
other tyrosine-regulated DAHP synthases as well. Sequence alignments with
phenylalanine-regulated DAHP synthases including Aro3p show a highly conserved
serine residue at this position. An exchange of glycine to serine and vice versa
leads to a complete change in the regulation pattern. Therefore the evolution of
these differently feedback-inhibited isoenzymes required gene duplication and a
single mutation within the internal extra element. Numerous additional amino
acid substitutions present in the contemporary isoenzymes are irrelevant for
regulation and occurred independently.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Aro4p structure of S. cerevisiae. (A) Topology
plot of DAHP synthase from S. cerevisiae. The  -strands and -strands and
 -helices of
the central -helices of
the central  / /  barrel are
shown in orange and yellow, respectively. Loops on the N- and
C-terminal sides of the barrel are in cyan and green,
respectively. The additional structural elements at the N
terminus and between helix barrel are
shown in orange and yellow, respectively. Loops on the N- and
C-terminal sides of the barrel are in cyan and green,
respectively. The additional structural elements at the N
terminus and between helix  5 and 5 and  -strand -strand  6 are also
shown in cyan. Residues involved in binding of PEP are marked in
green. Residues involved in regulation are marked in blue with
italic lettering for those identified by random screening and
normal lettering for those found by site-directed mutagenesis.
Red arrows mark the cutting points in Aro4p for the chimera
constructs (Fig. 1A). The dotted 6 are also
shown in cyan. Residues involved in binding of PEP are marked in
green. Residues involved in regulation are marked in blue with
italic lettering for those identified by random screening and
normal lettering for those found by site-directed mutagenesis.
Red arrows mark the cutting points in Aro4p for the chimera
constructs (Fig. 1A). The dotted  0 strand is
part of the second monomer of a dimer and interacts with the 0 strand is
part of the second monomer of a dimer and interacts with the
 6a and 6a and  6b. (B) The
crystal structure of DAHP synthase from S. cerevisiae in ribbon
presentation. The crystal structures contain a molecule of PEP
bound to the active site shown as space-filling models (red and
magenta). Secondary structure elements are color-coded as
described for A. Shown are views from the C-terminal face of the
central 6b. (B) The
crystal structure of DAHP synthase from S. cerevisiae in ribbon
presentation. The crystal structures contain a molecule of PEP
bound to the active site shown as space-filling models (red and
magenta). Secondary structure elements are color-coded as
described for A. Shown are views from the C-terminal face of the
central  barrel
(Left), side of the barrel (Center), and N-terminal face of the
barrel (Right). The programs MOLSCRIPT 2.1 (21) and RASTER3D
(22) were used for the presentation of DAHP synthase. barrel
(Left), side of the barrel (Center), and N-terminal face of the
barrel (Right). The programs MOLSCRIPT 2.1 (21) and RASTER3D
(22) were used for the presentation of DAHP synthase.
|
 |
Figure 5.
Fig. 5. Comparison of the putative effector-binding
cavity of the tyrosine- and phenylalanine-regulated DAHP
synthases. (A) Amino acid sequence alignment of a part of the
putative regulation cavity of the tyrosine- and
phenylalanine-regulated DAHP synthases of several organisms. C.
albicans, Candida albicans; A. nidulans, Aspergillus nidulans;
H. influenzae, Haemophilus influenzae; S. typhimurium,
Salmonella typhimurium. (B-D) Accessible surface plots of the
tyrosine-regulated DAHP synthase Aro4p from S. cerevisiae (S.c.,
B and C) and the phenylalanine-inhibited DAHP synthase AroG from
E. coli (E.c., D). Surface elements closer than 3 Å to
atoms belonging to the N-terminal extension or the inserted  sheet are
shown in cyan. Residues identified as playing a role in
regulation (blue) and for the specificity-related residue (red,
G226 in Aro4p and S211 in AroG) are indicated. (B) The
orientation is the same as described for Fig. 2B. (C and D) The
view is rotated by 40° around the vertical and 30°
around the horizontal axis with respect to Fig. 2B. The figure
was created with DINO (ref. 23 and www.dino3d.org). sheet are
shown in cyan. Residues identified as playing a role in
regulation (blue) and for the specificity-related residue (red,
G226 in Aro4p and S211 in AroG) are indicated. (B) The
orientation is the same as described for Fig. 2B. (C and D) The
view is rotated by 40° around the vertical and 30°
around the horizontal axis with respect to Fig. 2B. The figure
was created with DINO (ref. 23 and www.dino3d.org).
|
 |
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Tapas,
G.Kumar Patel,
S.Dhindwal,
and
S.Tomar
(2011).
In Silico sequence analysis and molecular modeling of the three-dimensional structure of DAHP synthase from Pseudomonas fragi.
|
| |
J Mol Model,
17,
621-631.
|
 |
|
|
|
|
 |
T.Casey,
P.S.Solomon,
S.Bringans,
K.C.Tan,
R.P.Oliver,
and
R.Lipscombe
(2010).
Quantitative proteomic analysis of G-protein signalling in Stagonospora nodorum using isobaric tags for relative and absolute quantification.
|
| |
Proteomics,
10,
38-47.
|
 |
|
|
|
|
 |
M.A.Luttik,
Z.Vuralhan,
E.Suir,
G.H.Braus,
J.T.Pronk,
and
J.M.Daran
(2008).
Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact.
|
| |
Metab Eng,
10,
141-153.
|
 |
|
|
|
|
 |
M.Calderón-Torres,
A.Peña,
and
P.E.Thomé
(2006).
DhARO4, an amino acid biosynthetic gene, is stimulated by high salinity in Debaryomyces hansenii.
|
| |
Yeast,
23,
725-734.
|
 |
|
|
|
|
 |
C.J.Webby,
J.S.Lott,
H.M.Baker,
E.N.Baker,
and
E.J.Parker
(2005).
Crystallization and preliminary X-ray crystallographic analysis of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
403-406.
|
 |
|
|
|
|
 |
K.Helmstaedt,
A.Strittmatter,
W.N.Lipscomb,
and
G.H.Braus
(2005).
Evolution of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase-encoding genes in the yeast Saccharomyces cerevisiae.
|
| |
Proc Natl Acad Sci U S A,
102,
9784-9789.
|
 |
|
|
|
|
 |
L.Kuepfer,
U.Sauer,
and
L.M.Blank
(2005).
Metabolic functions of duplicate genes in Saccharomyces cerevisiae.
|
| |
Genome Res,
15,
1421-1430.
|
 |
|
|
|
|
 |
M.S.Yousef,
W.A.Baase,
and
B.W.Matthews
(2004).
Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme.
|
| |
Proc Natl Acad Sci U S A,
101,
11583-11586.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

