 |
PDBsum entry 1he5

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biliverdin-ix beta reductase
|
PDB id
|
|

|

|
1he5
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Biliverdin-ix beta reductase
|
 |
|
Title:
|
 |
Human biliverdin ix beta reductase: NADP/lumichrome ternary complex
|
|
Structure:
|
 |
Biliverdin ix beta reductase. Chain: a. Synonym: flavin reductase (ec 1.6.99.1), NADPH-dependent diaphorase, NADPH-flavin reductase, biliverdin reductase b, green heme binding protein. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.50Å
|
R-factor:
|
0.148
|
R-free:
|
0.192
|
|
|
Authors:
|
 |
P.J.B.Pereira,S.Macedo-Ribeiro,A.Parraga,R.Perez-Luque,O.Cunningham, K.Darcy,T.J.Mantle,M.Coll
|
Key ref:

|
 |
P.J.Pereira
et al.
(2001).
Structure of human biliverdin IXbeta reductase, an early fetal bilirubin IXbeta producing enzyme.
Nat Struct Biol,
8,
215-220.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
19-Nov-00
|
Release date:
|
28-Feb-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P30043
(BLVRB_HUMAN) -
Flavin reductase (NADPH) from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
206 a.a.
205 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.3.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.5.1.30
- flavin reductase (NADPH).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
reduced riboflavin + NADP+ = riboflavin + NADPH + 2 H+
|
 |
 |
 |
 |
 |
reduced riboflavin
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NADP(+)
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
=
|
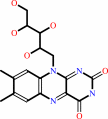 riboflavin
riboflavin
|
+
|
 NADPH
NADPH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.2.6.99.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
8:215-220
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of human biliverdin IXbeta reductase, an early fetal bilirubin IXbeta producing enzyme.
|
|
P.J.Pereira,
S.Macedo-Ribeiro,
A.Párraga,
R.Pérez-Luque,
O.Cunningham,
K.Darcy,
T.J.Mantle,
M.Coll.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Biliverdin IXbeta reductase (BVR-B) catalyzes the pyridine nucleotide-dependent
production of bilirubin-IXbeta, the major heme catabolite during early fetal
development. BVR-B displays a preference for biliverdin isomers without
propionates straddling the C10 position, in contrast to biliverdin IXalpha
reductase (BVR-A), the major form of BVR in adult human liver. In addition to
its tetrapyrrole clearance role in the fetus, BVR-B has flavin and ferric
reductase activities in the adult. We have solved the structure of human BVR-B
in complex with NADP+ at 1.15 A resolution. Human BVR-B is a monomer displaying
an alpha/beta dinucleotide binding fold. The structures of ternary complexes
with mesobiliverdin IValpha, biliverdin IXalpha, FMN and lumichrome show that
human BVR-B has a single substrate binding site, to which substrates and
inhibitors bind primarily through hydrophobic interactions, explaining its broad
specificity. The reducible atom of both biliverdin and flavin substrates lies
above the reactive C4 of the cofactor, an appropriate position for direct
hydride transfer. BVR-B discriminates against the biliverdin IXalpha isomer
through steric hindrance at the bilatriene side chain binding pockets. The
structure also explains the enzyme's preference for NADP(H) and its B-face
stereospecificity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Human BVR-B binds specifically to NADP. a, Stereo
view of the NADP+ cofactor binding site. Putative hydrogen bonds
are shown as dotted green lines, and solvent molecules as red
spheres. The atom color code is the same as in Fig. 1b. The 1.15
Å 2F[o] - F[c] electron density map for the cofactor, contoured
at 1.5  ,
is shown in blue. Residues interacting with NADP+ are numbered.
b, Solid surface representation of human BVR-B in complex with
NADP+ showing the wide substrate binding site adjacent to the
cofactor. Electrostatic surface potentials are contoured from
-10 (red) to 10 (blue) k[B]T e^-1. The cofactor carbon atoms are
shown in white, phosphorous in yellow, nitrogens in blue and
oxygens in red. ,
is shown in blue. Residues interacting with NADP+ are numbered.
b, Solid surface representation of human BVR-B in complex with
NADP+ showing the wide substrate binding site adjacent to the
cofactor. Electrostatic surface potentials are contoured from
-10 (red) to 10 (blue) k[B]T e^-1. The cofactor carbon atoms are
shown in white, phosphorous in yellow, nitrogens in blue and
oxygens in red.
|
 |
Figure 3.
Figure 3. Structural formulas of some human BVR-B substrates and
inhibitors. a, FMN is a BVR-B substrate, lumichrome an
inhibitor. b, Tetrapyrrole biliverdin IX  ,
mesobiliverdin IV ,
mesobiliverdin IV  and
12-ethyl-13-methyl-mesobiliverdin IV and
12-ethyl-13-methyl-mesobiliverdin IV  are
substrates; biliverdin IX are
substrates; biliverdin IX  is
an inhibitor. The formation of a bilirubin isomer results from
the reduction of C10 of the corresponding biliverdin (numbering
as indicated for biliverdin IX is
an inhibitor. The formation of a bilirubin isomer results from
the reduction of C10 of the corresponding biliverdin (numbering
as indicated for biliverdin IX  ,
which is conserved for the bilatriene skeleton in all other
isomers). The ring nomenclature for all ,
which is conserved for the bilatriene skeleton in all other
isomers). The ring nomenclature for all  -isomers
is that shown for biliverdin IX -isomers
is that shown for biliverdin IX  . .
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2001,
8,
215-220)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Hassaninasab,
Y.Hashimoto,
K.Tomita-Yokotani,
and
M.Kobayashi
(2011).
Discovery of the curcumin metabolic pathway involving a unique enzyme in an intestinal microorganism.
|
| |
Proc Natl Acad Sci U S A,
108,
6615-6620.
|
 |
|
|
|
|
 |
D.Méndez,
M.Linares,
A.Diez,
A.Puyet,
and
J.M.Bautista
(2011).
Stress response and cytoskeletal proteins involved in erythrocyte membrane remodeling upon Plasmodium falciparum invasion are differentially carbonylated in G6PD A- deficiency.
|
| |
Free Radic Biol Med,
50,
1305-1313.
|
 |
|
|
|
|
 |
H.Chen,
J.Feng,
O.Kweon,
H.Xu,
and
C.E.Cerniglia
(2010).
Identification and molecular characterization of a novel flavin-free NADPH preferred azoreductase encoded by azoB in Pigmentiphaga kullae K24.
|
| |
BMC Biochem,
11,
13.
|
 |
|
|
|
|
 |
E.M.Franklin,
S.Browne,
A.M.Horan,
K.Inomata,
M.A.Hammam,
H.Kinoshita,
T.Lamparter,
G.Golfis,
and
T.J.Mantle
(2009).
The use of synthetic linear tetrapyrroles to probe the verdin sites of human biliverdin-IXalpha reductase and human biliverdin-IXbeta reductase.
|
| |
FEBS J,
276,
4405-4413.
|
 |
|
|
|
|
 |
J.M.Hayes,
and
T.J.Mantle
(2009).
The effect of pH on the initial rate kinetics of the dimeric biliverdin-IXalpha reductase from the cyanobacterium Synechocystis PCC6803.
|
| |
FEBS J,
276,
4414-4425.
|
 |
|
|
|
|
 |
A.K.Sendamarai,
R.S.Ohgami,
M.D.Fleming,
and
C.M.Lawrence
(2008).
Structure of the membrane proximal oxidoreductase domain of human Steap3, the dominant ferrireductase of the erythroid transferrin cycle.
|
| |
Proc Natl Acad Sci U S A,
105,
7410-7415.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.L.Kavanagh,
H.Jörnvall,
B.Persson,
and
U.Oppermann
(2008).
Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes.
|
| |
Cell Mol Life Sci,
65,
3895-3906.
|
 |
|
|
|
|
 |
J.D.King,
N.J.Harmer,
A.Preston,
C.M.Palmer,
M.Rejzek,
R.A.Field,
T.L.Blundell,
and
D.J.Maskell
(2007).
Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
|
| |
J Mol Biol,
374,
749-763.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.S.Jang,
N.Y.Kang,
K.S.Kim,
C.H.Kim,
J.H.Lee,
and
Y.C.Lee
(2007).
Mutational analysis of NADH-binding residues in triphenylmethane reductase from Citrobacter sp. strain KCTC 18061P.
|
| |
FEMS Microbiol Lett,
271,
78-82.
|
 |
|
|
|
|
 |
M.Unno,
T.Matsui,
and
M.Ikeda-Saito
(2007).
Structure and catalytic mechanism of heme oxygenase.
|
| |
Nat Prod Rep,
24,
553-570.
|
 |
|
|
|
|
 |
B.Youn,
R.Camacho,
S.G.Moinuddin,
C.Lee,
L.B.Davin,
N.G.Lewis,
and
C.Kang
(2006).
Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4.
|
| |
Org Biomol Chem,
4,
1687-1697.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Youn,
S.J.Kim,
S.G.Moinuddin,
C.Lee,
D.L.Bedgar,
A.R.Harper,
L.B.Davin,
N.G.Lewis,
and
C.Kang
(2006).
Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
|
| |
J Biol Chem,
281,
40076-40088.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Youn,
S.G.Moinuddin,
L.B.Davin,
N.G.Lewis,
and
C.Kang
(2005).
Crystal structures of apo-form and binary/ternary complexes of Podophyllum secoisolariciresinol dehydrogenase, an enzyme involved in formation of health-protecting and plant defense lignans.
|
| |
J Biol Chem,
280,
12917-12926.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.El Omari,
L.E.Bird,
C.E.Nichols,
J.Ren,
and
D.K.Stammers
(2005).
Crystal structure of CC3 (TIP30): implications for its role as a tumor suppressor.
|
| |
J Biol Chem,
280,
18229-18236.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.W.Schultz,
L.Liu,
M.Cegielski,
and
J.W.Hastings
(2005).
Crystal structure of a pH-regulated luciferase catalyzing the bioluminescent oxidation of an open tetrapyrrole.
|
| |
Proc Natl Acad Sci U S A,
102,
1378-1383.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Kukimoto-Niino,
K.Murayama,
M.Kato-Murayama,
M.Idaka,
Y.Bessho,
A.Tatsuguchi,
R.Ushikoshi-Nakayama,
T.Terada,
S.Kuramitsu,
M.Shirouzu,
and
S.Yokoyama
(2004).
Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8.
|
| |
Protein Sci,
13,
3038-3042.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Sarkhel,
and
G.R.Desiraju
(2004).
N-H...O, O-H...O, and C-H...O hydrogen bonds in protein-ligand complexes: strong and weak interactions in molecular recognition.
|
| |
Proteins,
54,
247-259.
|
 |
|
|
|
|
 |
J.Maclean,
and
S.Ali
(2003).
The structure of chorismate synthase reveals a novel flavin binding site fundamental to a unique chemical reaction.
|
| |
Structure,
11,
1499-1511.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Wang,
and
P.R.de Montellano
(2003).
The binding sites on human heme oxygenase-1 for cytochrome p450 reductase and biliverdin reductase.
|
| |
J Biol Chem,
278,
20069-20076.
|
 |
|
|
|
|
 |
M.Sugishima,
H.Sakamoto,
Y.Higashimoto,
M.Noguchi,
and
K.Fukuyama
(2003).
Crystal structure of rat heme oxygenase-1 in complex with biliverdin-iron chelate. Conformational change of the distal helix during the heme cleavage reaction.
|
| |
J Biol Chem,
278,
32352-32358.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Min,
H.Kasahara,
D.L.Bedgar,
B.Youn,
P.K.Lawrence,
D.R.Gang,
S.C.Halls,
H.Park,
J.L.Hilsenbeck,
L.B.Davin,
N.G.Lewis,
C.Kang,
and
N.G.Lewis
(2003).
Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases.
|
| |
J Biol Chem,
278,
50714-50723.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.A.Bottoms,
P.E.Smith,
and
J.J.Tanner
(2002).
A structurally conserved water molecule in Rossmann dinucleotide-binding domains.
|
| |
Protein Sci,
11,
2125-2137.
|
 |
|
|
|
|
 |
D.A.Greenberg
(2002).
The jaundice of the cell.
|
| |
Proc Natl Acad Sci U S A,
99,
15837-15839.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

