 |
PDBsum entry 1hdh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Arylsulfatase from pseudomonas aeruginosa
|
|
Structure:
|
 |
Arylsulfatase. Chain: a, b. Synonym: as,aryl-sulfate sulphohydrolase. Engineered: yes
|
|
Source:
|
 |
Pseudomonas aeruginosa. Organism_taxid: 287. Gene: atsa, pa0183. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.30Å
|
R-factor:
|
0.200
|
R-free:
|
0.229
|
|
|
Authors:
|
 |
I.Boltes,H.Czapinska,A.Kahnert,R.Von Buelow,T.Dirks,B.Schmidt,K.Von Figura,M.A.Kertesz,I.Uson
|
Key ref:

|
 |
I.Boltes
et al.
(2001).
1.3 A structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family.
Structure,
9,
483-491.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
16-Nov-00
|
Release date:
|
15-Nov-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P51691
(ARS_PSEAE) -
Arylsulfatase from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
536 a.a.
525 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.6.1
- arylsulfatase (type I).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an aryl sulfate + H2O = a phenol + sulfate + H+
|
 |
 |
 |
 |
 |
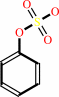 aryl sulfate
aryl sulfate
|
+
|
H2O
|
=
|
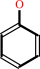 phenol
phenol
|
+
|
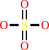 sulfate
sulfate
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
9:483-491
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
1.3 A structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family.
|
|
I.Boltes,
H.Czapinska,
A.Kahnert,
R.von Bülow,
T.Dierks,
B.Schmidt,
K.von Figura,
M.A.Kertesz,
I.Usón.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Sulfatases constitute a family of enzymes with a highly conserved
active site region including a Calpha-formylglycine that is posttranslationally
generated by the oxidation of a conserved cysteine or serine residue. The
crystal structures of two human arylsulfatases, ASA and ASB, along with ASA
mutants and their complexes led to different proposals for the catalytic
mechanism in the hydrolysis of sulfate esters. RESULTS: The crystal structure of
a bacterial sulfatase from Pseudomonas aeruginosa (PAS) has been determined at
1.3 A. Fold and active site region are strikingly similar to those of the known
human sulfatases. The structure allows a precise determination of the active
site region, unequivocally showing the presence of a Calpha-formylglycine
hydrate as the key catalytic residue. Furthermore, the cation located in the
active site is unambiguously characterized as calcium by both its B value and
the geometry of its coordination sphere. The active site contains a
noncovalently bonded sulfate that occupies the same position as the one in
para-nitrocatecholsulfate in previously studied ASA complexes. CONCLUSIONS: The
structure of PAS shows that the resting state of the key catalytic residue in
sulfatases is a formylglycine hydrate. These structural data establish a
mechanism for sulfate ester cleavage involving an aldehyde hydrate as the
functional group that initiates the reaction through a nucleophilic attack on
the sulfur atom in the substrate. The alcohol is eliminated from a reaction
intermediate containing pentacoordinated sulfur. Subsequent elimination of the
sulfate regenerates the aldehyde, which is again hydrated. The metal cation
involved in stabilizing the charge and anchoring the substrate during catalysis
is established as calcium.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 5.
Figure 5. A scheme of the Proposed Catalytic Mechanism

|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2001,
9,
483-491)
copyright 2001.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Lu,
M.E.Wörmann,
X.Zhang,
O.Schneewind,
A.Gründling,
and
P.S.Freemont
(2009).
Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS.
|
| |
Proc Natl Acad Sci U S A,
106,
1584-1589.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.R.Myette,
V.Soundararajan,
J.Behr,
Z.Shriver,
R.Raman,
and
R.Sasisekharan
(2009).
Heparin/heparan sulfate N-sulfamidase from Flavobacterium heparinum: structural and biochemical investigation of catalytic nitrogen-sulfur bond cleavage.
|
| |
J Biol Chem,
284,
35189-35200.
|
 |
|
|
|
|
 |
J.R.Myette,
V.Soundararajan,
Z.Shriver,
R.Raman,
and
R.Sasisekharan
(2009).
Heparin/heparan sulfate 6-O-sulfatase from Flavobacterium heparinum: integrated structural and biochemical investigation of enzyme active site and substrate specificity.
|
| |
J Biol Chem,
284,
35177-35188.
|
 |
|
|
|
|
 |
K.Schirner,
J.Marles-Wright,
R.J.Lewis,
and
J.Errington
(2009).
Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis.
|
| |
EMBO J,
28,
830-842.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.A.Frese,
and
T.Dierks
(2009).
Formylglycine aldehyde Tag--protein engineering through a novel post-translational modification.
|
| |
Chembiochem,
10,
425-427.
|
 |
|
|
|
|
 |
M.Schenk,
C.A.Koppisetty,
D.C.Santos,
E.Carmona,
S.Bhatia,
P.G.Nyholm,
and
N.Tanphaichitr
(2009).
Interaction of arylsulfatase-A (ASA) with its natural sulfoglycolipid substrates: a computational and site-directed mutagenesis study.
|
| |
Glycoconj J,
26,
1029-1045.
|
 |
|
|
|
|
 |
J.P.Lai,
J.R.Thompson,
D.S.Sandhu,
and
L.R.Roberts
(2008).
Heparin-degrading sulfatases in hepatocellular carcinoma: roles in pathogenesis and therapy targets.
|
| |
Future Oncol,
4,
803-814.
|
 |
|
|
|
|
 |
M.Mariappan,
K.Radhakrishnan,
T.Dierks,
B.Schmidt,
and
K.von Figura
(2008).
ERp44 mediates a thiol-independent retention of formylglycine-generating enzyme in the endoplasmic reticulum.
|
| |
J Biol Chem,
283,
6375-6383.
|
 |
|
|
|
|
 |
P.Bojarová,
E.Denehy,
I.Walker,
K.Loft,
D.P.De Souza,
L.W.Woo,
B.V.Potter,
M.J.McConville,
and
S.J.Williams
(2008).
Direct evidence for ArO-S bond cleavage upon inactivation of Pseudomonas aeruginosa arylsulfatase by aryl sulfamates.
|
| |
Chembiochem,
9,
613-623.
|
 |
|
|
|
|
 |
P.Bojarová,
and
S.J.Williams
(2008).
Sulfotransferases, sulfatases and formylglycine-generating enzymes: a sulfation fascination.
|
| |
Curr Opin Chem Biol,
12,
573-581.
|
 |
|
|
|
|
 |
S.L.Gande,
M.Mariappan,
B.Schmidt,
T.H.Pringle,
K.von Figura,
and
T.Dierks
(2008).
Paralog of the formylglycine-generating enzyme--retention in the endoplasmic reticulum by canonical and noncanonical signals.
|
| |
FEBS J,
275,
1118-1130.
|
 |
|
|
|
|
 |
T.L.Grove,
K.H.Lee,
J.St Clair,
C.Krebs,
and
S.J.Booker
(2008).
In vitro characterization of AtsB, a radical SAM formylglycine-generating enzyme that contains three [4Fe-4S] clusters.
|
| |
Biochemistry,
47,
7523-7538.
|
 |
|
|
|
|
 |
E.Zito,
M.Buono,
S.Pepe,
C.Settembre,
I.Annunziata,
E.M.Surace,
T.Dierks,
M.Monti,
M.Cozzolino,
P.Pucci,
A.Ballabio,
and
M.P.Cosma
(2007).
Sulfatase modifying factor 1 trafficking through the cells: from endoplasmic reticulum to the endoplasmic reticulum.
|
| |
EMBO J,
26,
2443-2453.
|
 |
|
|
|
|
 |
P.Gadler,
and
K.Faber
(2007).
New enzymes for biotransformations: microbial alkyl sulfatases displaying stereo- and enantioselectivity.
|
| |
Trends Biotechnol,
25,
83-88.
|
 |
|
|
|
|
 |
J.Rush,
and
C.R.Bertozzi
(2006).
An alpha-formylglycine building block for fmoc-based solid-phase peptide synthesis.
|
| |
Org Lett,
8,
131-134.
|
 |
|
|
|
|
 |
O.Berteau,
A.Guillot,
A.Benjdia,
and
S.Rabot
(2006).
A new type of bacterial sulfatase reveals a novel maturation pathway in prokaryotes.
|
| |
J Biol Chem,
281,
22464-22470.
|
 |
|
|
|
|
 |
S.R.Hanson,
L.J.Whalen,
and
C.H.Wong
(2006).
Synthesis and evaluation of general mechanism-based inhibitors of sulfatases based on (difluoro)methyl phenyl sulfate and cyclic phenyl sulfamate motifs.
|
| |
Bioorg Med Chem,
14,
8386-8395.
|
 |
|
|
|
|
 |
A.Preusser-Kunze,
M.Mariappan,
B.Schmidt,
S.L.Gande,
K.Mutenda,
D.Wenzel,
K.von Figura,
and
T.Dierks
(2005).
Molecular characterization of the human Calpha-formylglycine-generating enzyme.
|
| |
J Biol Chem,
280,
14900-14910.
|
 |
|
|
|
|
 |
D.Roeser,
A.Dickmanns,
K.Gasow,
and
M.G.Rudolph
(2005).
De novo calcium/sulfur SAD phasing of the human formylglycine-generating enzyme using in-house data.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1057-1066.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Mariappan,
A.Preusser-Kunze,
M.Balleininger,
N.Eiselt,
B.Schmidt,
S.L.Gande,
D.Wenzel,
T.Dierks,
and
K.von Figura
(2005).
Expression, localization, structural, and functional characterization of pFGE, the paralog of the Calpha-formylglycine-generating enzyme.
|
| |
J Biol Chem,
280,
15173-15179.
|
 |
|
|
|
|
 |
S.R.Wallner,
B.M.Nestl,
and
K.Faber
(2005).
Highly enantioselective stereo-inverting sec-alkylsulfatase activity of hyperthermophilic Archaea.
|
| |
Org Biomol Chem,
3,
2652-2656.
|
 |
|
|
|
|
 |
S.R.Wallner,
M.Bauer,
C.Würdemann,
P.Wecker,
F.O.Glöckner,
and
K.Faber
(2005).
Highly enantioselective sec-alkyl sulfatase activity of the marine planctomycete Rhodopirellula baltica shows retention of configuration.
|
| |
Angew Chem Int Ed Engl,
44,
6381-6384.
|
 |
|
|
|
|
 |
Q.Fang,
J.Peng,
and
T.Dierks
(2004).
Post-translational formylglycine modification of bacterial sulfatases by the radical S-adenosylmethionine protein AtsB.
|
| |
J Biol Chem,
279,
14570-14578.
|
 |
|
|
|
|
 |
S.R.Hanson,
M.D.Best,
and
C.H.Wong
(2004).
Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility.
|
| |
Angew Chem Int Ed Engl,
43,
5736-5763.
|
 |
|
|
|
|
 |
C.Marquordt,
Q.Fang,
E.Will,
J.Peng,
K.von Figura,
and
T.Dierks
(2003).
Posttranslational modification of serine to formylglycine in bacterial sulfatases. Recognition of the modification motif by the iron-sulfur protein AtsB.
|
| |
J Biol Chem,
278,
2212-2218.
|
 |
|
|
|
|
 |
J.Peng,
B.Schmidt,
K.von Figura,
and
T.Dierks
(2003).
Identification of formylglycine in sulfatases by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
|
| |
J Mass Spectrom,
38,
80-86.
|
 |
|
|
|
|
 |
K.Y.Cheng,
E.D.Lowe,
J.Sinclair,
E.A.Nigg,
and
L.N.Johnson
(2003).
The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex.
|
| |
EMBO J,
22,
5757-5768.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Steinbacher,
S.Schiffmann,
G.Richter,
R.Huber,
A.Bacher,
and
M.Fischer
(2003).
Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with divalent metal ions and the substrate ribulose 5-phosphate: implications for the catalytic mechanism.
|
| |
J Biol Chem,
278,
42256-42265.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Dierks,
B.Schmidt,
L.V.Borissenko,
J.Peng,
A.Preusser,
M.Mariappan,
and
K.von Figura
(2003).
Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme.
|
| |
Cell,
113,
435-444.
|
 |
|
|
|
|
 |
J.D.Mougous,
R.E.Green,
S.J.Williams,
S.E.Brenner,
and
C.R.Bertozzi
(2002).
Sulfotransferases and sulfatases in mycobacteria.
|
| |
Chem Biol,
9,
767-776.
|
 |
|
|
|
|
 |
J.Fey,
M.Balleininger,
L.V.Borissenko,
B.Schmidt,
K.von Figura,
and
T.Dierks
(2001).
Characterization of posttranslational formylglycine formation by luminal components of the endoplasmic reticulum.
|
| |
J Biol Chem,
276,
47021-47028.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

