 |
PDBsum entry 1g0d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.3.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.3.2.13
- protein-glutamine gamma-glutamyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutaminyl-[protein] + L-lysyl-[protein] = [protein]-L-lysyl-N6-5- L-glutamyl-[protein] + NH4+
|
 |
 |
 |
 |
 |
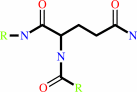 protein-L-glutamine
protein-L-glutamine
|
+
|
protein-L-lysine
|
=
|
protein with an N(6)- (gamma-glutamyl)-L-lysine cross-link
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Ca(2+)
|
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.4.-.-
|
|
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.3.5.1.44
- protein-glutamine glutaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutaminyl-[protein] + H2O = L-glutamyl-[protein] + NH4+
|
 |
 |
 |
 |
 |
 Protein L-glutamine
Protein L-glutamine
|
+
|
H(2)O
|
=
|
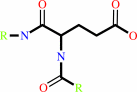 protein L-glutamate
protein L-glutamate
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
276:12055-12059
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of red sea bream transglutaminase.
|
|
K.Noguchi,
K.Ishikawa,
Yokoyama Ki,
T.Ohtsuka,
N.Nio,
E.Suzuki.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the tissue-type transglutaminase from red sea bream
liver (fish-derived transglutaminase, FTG) has been determined at 2.5-A
resolution using the molecular replacement method, based on the crystal
structure of human blood coagulation factor XIII, which is a transglutaminase
zymogen. The model contains 666 residues of a total of 695 residues, 382 water
molecules, and 1 sulfate ion. FTG consists of four domains, and its overall and
active site structures are similar to those of human factor XIII. However,
significant structural differences are observed in both the acyl donor and acyl
acceptor binding sites, which account for the difference in substrate
preferences. The active site of the enzyme is inaccessible to the solvent,
because the catalytic Cys-272 hydrogen-bonds to Tyr-515, which is thought to be
displaced upon acyl donor binding to FTG. It is postulated that the binding of
an inappropriate substrate to FTG would lead to inactivation of the enzyme
because of the formation of a new disulfide bridge between Cys-272 and the
adjacent Cys-333 immediately after the displacement of Tyr-515. Considering the
mutational studies previously reported on the tissue-type transglutaminases, we
propose that Cys-333 and Tyr-515 are important in strictly controlling the
enzymatic activity of FTG.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Schematic ribbon drawings of FTG (left) and human
factor XIII (right). Helices and sheets are colored red and
blue, respectively. Both TGases consist of four domains:  -sandwich,
core, barrel 1, and barrel 2 from the N terminus. Human factor
XIII has an activation peptide (green) at the N terminus. The
active site in each TGase is marked with a yellow asterisk. This
figure was produced using MOLSCRIPT (36). -sandwich,
core, barrel 1, and barrel 2 from the N terminus. Human factor
XIII has an activation peptide (green) at the N terminus. The
active site in each TGase is marked with a yellow asterisk. This
figure was produced using MOLSCRIPT (36).
|
 |
Figure 2.
Fig. 2. Structural comparison of FTG with human factor
XIII. Stereoviews of the C  traces of
FTG and human factor XIII are shown. The superposition was done
using QUANTA97. FTG, human factor XIII, and the activation
peptide are colored blue, red, and green, respectively. The
active site in each TGase is marked with a white asterisk. traces of
FTG and human factor XIII are shown. The superposition was done
using QUANTA97. FTG, human factor XIII, and the activation
peptide are colored blue, red, and green, respectively. The
active site in each TGase is marked with a white asterisk.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2001,
276,
12055-12059)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.S.Bhaskaran,
and
C.E.Stebbins
(2012).
Structure of the catalytic domain of the Salmonella virulence factor SseI.
|
| |
Acta Crystallogr D Biol Crystallogr,
68,
1613-1621.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.A.Wilson,
and
M.Ho
(2010).
Recent insights into Pasteurella multocida toxin and other G-protein-modulating bacterial toxins.
|
| |
Future Microbiol,
5,
1185-1201.
|
 |
|
|
|
|
 |
H.Kumeta,
N.Miwa,
K.Ogura,
Y.Kai,
T.Mizukoshi,
N.Shimba,
E.Suzuki,
and
F.Inagaki
(2010).
The NMR structure of protein-glutaminase from Chryseobacterium proteolyticum.
|
| |
J Biomol NMR,
46,
251-255.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
U.Tagami,
N.Shimba,
M.Nakamura,
K.Yokoyama,
E.Suzuki,
and
T.Hirokawa
(2009).
Substrate specificity of microbial transglutaminase as revealed by three-dimensional docking simulation and mutagenesis.
|
| |
Protein Eng Des Sel,
22,
747-752.
|
 |
|
|
|
|
 |
D.M.Pinkas,
P.Strop,
A.T.Brunger,
and
C.Khosla
(2007).
Transglutaminase 2 undergoes a large conformational change upon activation.
|
| |
PLoS Biol,
5,
e327.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.E.Begg,
L.Carrington,
P.H.Stokes,
J.M.Matthews,
M.A.Wouters,
A.Husain,
L.Lorand,
S.E.Iismaa,
and
R.M.Graham
(2006).
Mechanism of allosteric regulation of transglutaminase 2 by GTP.
|
| |
Proc Natl Acad Sci U S A,
103,
19683-19688.
|
 |
|
|
|
|
 |
R.A.Chica,
P.Gagnon,
J.W.Keillor,
and
J.N.Pelletier
(2004).
Tissue transglutaminase acylation: Proposed role of conserved active site Tyr and Trp residues revealed by molecular modeling of peptide substrate binding.
|
| |
Protein Sci,
13,
979-991.
|
 |
|
|
|
|
 |
A.Kon,
H.Takeda,
H.Sasaki,
K.Yoneda,
K.Nomura,
B.Ahvazi,
P.M.Steinert,
K.Hanada,
and
I.Hashimoto
(2003).
Novel transglutaminase 1 gene mutations (R348X/Y365D) in a Japanese family with lamellar ichthyosis.
|
| |
J Invest Dermatol,
120,
170-172.
|
 |
|
|
|
|
 |
L.Lorand,
and
R.M.Graham
(2003).
Transglutaminases: crosslinking enzymes with pleiotropic functions.
|
| |
Nat Rev Mol Cell Biol,
4,
140-156.
|
 |
|
|
|
|
 |
M.Date,
K.Yokoyama,
Y.Umezawa,
H.Matsui,
and
Y.Kikuchi
(2003).
Production of native-type Streptoverticillium mobaraense transglutaminase in Corynebacterium glutamicum.
|
| |
Appl Environ Microbiol,
69,
3011-3014.
|
 |
|
|
|
|
 |
S.E.Iismaa,
S.Holman,
M.A.Wouters,
L.Lorand,
R.M.Graham,
and
A.Husain
(2003).
Evolutionary specialization of a tryptophan indole group for transition-state stabilization by eukaryotic transglutaminases.
|
| |
Proc Natl Acad Sci U S A,
100,
12636-12641.
|
 |
|
|
|
|
 |
Y.Kikuchi,
M.Date,
K.Yokoyama,
Y.Umezawa,
and
H.Matsui
(2003).
Secretion of active-form Streptoverticillium mobaraense transglutaminase by Corynebacterium glutamicum: processing of the pro-transglutaminase by a cosecreted subtilisin-Like protease from Streptomyces albogriseolus.
|
| |
Appl Environ Microbiol,
69,
358-366.
|
 |
|
|
|
|
 |
B.Ahvazi,
H.C.Kim,
S.H.Kee,
Z.Nemes,
and
P.M.Steinert
(2002).
Three-dimensional structure of the human transglutaminase 3 enzyme: binding of calcium ions changes structure for activation.
|
| |
EMBO J,
21,
2055-2067.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Brunner,
S.Rosahl,
J.Lee,
J.J.Rudd,
C.Geiler,
S.Kauppinen,
G.Rasmussen,
D.Scheel,
and
T.Nürnberger
(2002).
Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases.
|
| |
EMBO J,
21,
6681-6688.
|
 |
|
|
|
|
 |
L.Fesus,
and
M.Piacentini
(2002).
Transglutaminase 2: an enigmatic enzyme with diverse functions.
|
| |
Trends Biochem Sci,
27,
534-539.
|
 |
|
|
|
|
 |
S.N.Murthy,
S.Iismaa,
G.Begg,
D.M.Freymann,
R.M.Graham,
and
L.Lorand
(2002).
Conserved tryptophan in the core domain of transglutaminase is essential for catalytic activity.
|
| |
Proc Natl Acad Sci U S A,
99,
2738-2742.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

