 |
PDBsum entry 1fp3

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.3.8
- N-acylglucosamine 2-epimerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acyl-D-glucosamine = an N-acyl-D-mannosamine
|
 |
 |
 |
 |
 |
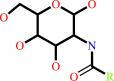 N-acyl-D-glucosamine
N-acyl-D-glucosamine
|
=
|
 N-acyl-D-mannosamine
N-acyl-D-mannosamine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
303:733-744
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of N-acyl-D-glucosamine 2-epimerase from porcine kidney at 2.0 A resolution.
|
|
T.Itoh,
B.Mikami,
I.Maru,
Y.Ohta,
W.Hashimoto,
K.Murata.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The X-ray crystallographic structure of N-acyl-d-glucosamine 2-epimerase (AGE)
from porcine kidney, which has been identified to be a renin-binding protein
(RnBP), was determined by the multiple isomorphous replacement method and
refined at 2.0 A resolution with a final R-factor of 16.9 % for 15 to 2.0 A
resolution data. The refined structure of AGE comprised 804 amino acid residues
(one dimer) and 145 water molecules. The dimer of AGE had an asymmetric unit
with approximate dimensions 46 Ax48 Ax96 A. The AGE monomer is composed of an
alpha(6)/alpha(6)-barrel, the structure of which is found in glucoamylase and
cellulase. One side of the AGE alpha(6)/alpha(6)-barrel structure comprises long
loops containing five short beta-sheets, and contributes to the formation of a
deep cleft shaped like a funnel. The putative active-site pocket and a possible
binding site for the substrate N-acetyl-d-glucosamine (GlcNAc) were found in the
cleft. The other side of the alpha(6)/alpha(6)-barrel comprises short loops and
contributes to the dimer formation. At the dimer interface, which is composed of
the short loops and alpha-helices of the subunits, five strong ion-pair
interactions were observed, which play a major role in the dimer assembly. This
completely ruled out the previously accepted hypothesis that the formation of
the RnBP homodimer and RnBP-renin heterodimer requires the leucine zipper motif
present in RnBP.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 6.
Figure 6. Interface of the AGE dimer. The structure is
represented as a pink and cyan molecular tube model of AGE
subunits A and B, respectively. Residues buried in the dimer
interface are presented in red (subunit A) and blue (subunit B).
The side-chains of residues forming hydrogen bonds or van der
Waals contacts are also colored. The broken line in the Figure
is the non-crystallographic 2-fold axis of the AGE dimer. The
Figure was drawn using the program GRASP [Nicholls et al 1991].
|
 |
Figure 7.
Figure 7. Structural comparison of a-helices within the
a/a-barrel structures of AGE (red), glucoamylase (cyan),
cellulase (yellow), and alginate lyase A1-III (green). The
superimposed results are shown as a schematic view in C^a-traces
of the a-helices. The coordinates of glucoamylase (1DOG),
cellulase (1CEM), and alginate lyase A1-III (1QAZ) were taken
from the RCSB Protein Data Bank [Berman et al 2000]. The Figure
was prepared using MOLSCRIPT [Kraulis 1991] and RASTER3D [Merrit
and Murphy 1994].
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
303,
733-744)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Brigham,
R.Caughlan,
R.Gallegos,
M.B.Dallas,
V.G.Godoy,
and
M.H.Malamy
(2009).
Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase.
|
| |
J Bacteriol,
191,
3629-3638.
|
 |
|
|
|
|
 |
S.Ito,
S.Hamada,
H.Ito,
H.Matsui,
T.Ozawa,
H.Taguchi,
and
S.Ito
(2009).
Site-directed mutagenesis of possible catalytic residues of cellobiose 2-epimerase from Ruminococcus albus.
|
| |
Biotechnol Lett,
31,
1065-1071.
|
 |
|
|
|
|
 |
T.Senoura,
H.Taguchi,
S.Ito,
S.Hamada,
H.Matsui,
S.Fukiya,
A.Yokota,
J.Watanabe,
J.Wasaki,
and
S.Ito
(2009).
Identification of the cellobiose 2-epimerase gene in the genome of Bacteroides fragilis NCTC 9343.
|
| |
Biosci Biotechnol Biochem,
73,
400-406.
|
 |
|
|
|
|
 |
Y.Maruyama,
Y.Nakamichi,
T.Itoh,
B.Mikami,
W.Hashimoto,
and
K.Murata
(2009).
Substrate specificity of streptococcal unsaturated glucuronyl hydrolases for sulfated glycosaminoglycan.
|
| |
J Biol Chem,
284,
18059-18069.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Murata,
S.Kawai,
B.Mikami,
and
W.Hashimoto
(2008).
Superchannel of bacteria: biological significance and new horizons.
|
| |
Biosci Biotechnol Biochem,
72,
265-277.
|
 |
|
|
|
|
 |
W.Hashimoto,
K.Momma,
Y.Maruyama,
M.Yamasaki,
B.Mikami,
and
K.Murata
(2005).
Structure and function of bacterial super-biosystem responsible for import and depolymerization of macromolecules.
|
| |
Biosci Biotechnol Biochem,
69,
673-692.
|
 |
|
|
|
|
 |
T.Itoh,
S.Akao,
W.Hashimoto,
B.Mikami,
and
K.Murata
(2004).
Crystal structure of unsaturated glucuronyl hydrolase, responsible for the degradation of glycosaminoglycan, from Bacillus sp. GL1 at 1.8 A resolution.
|
| |
J Biol Chem,
279,
31804-31812.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Hashimoto,
M.Yamasaki,
T.Itoh,
K.Momma,
B.Mikami,
and
K.Murata
(2004).
Super-channel in bacteria: structural and functional aspects of a novel biosystem for the import and depolymerization of macromolecules.
|
| |
J Biosci Bioeng,
98,
399-413.
|
 |
|
|
|
|
 |
W.Hashimoto,
H.Nankai,
B.Mikami,
and
K.Murata
(2003).
Crystal structure of Bacillus sp. GL1 xanthan lyase, which acts on the side chains of xanthan.
|
| |
J Biol Chem,
278,
7663-7673.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Maru,
J.Ohnishi,
Y.Ohta,
and
Y.Tsukada
(2002).
Why is sialic acid attracting interest now? Complete enzymatic synthesis of sialic acid with N-acylglucosamine 2-epimerase.
|
| |
J Biosci Bioeng,
93,
258-265.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

