 |
PDBsum entry 1fj2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.2.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.2.22
- palmitoyl-protein hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-hexadecanoyl-L-cysteinyl-[protein] + H2O = L-cysteinyl-[protein] + hexadecanoate + H+
|
 |
 |
 |
 |
 |
S-hexadecanoyl-L-cysteinyl-[protein]
|
+
|
H2O
|
=
|
L-cysteinyl-[protein]
|
+
|
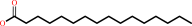 hexadecanoate
hexadecanoate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
8:1137-1146
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A.
|
|
Y.Devedjiev,
Z.Dauter,
S.R.Kuznetsov,
T.L.Jones,
Z.S.Derewenda.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Many proteins undergo posttranslational modifications involving
covalent attachment of lipid groups. Among them is palmitoylation, a dynamic,
reversible process that affects trimeric G proteins and Ras and constitutes a
regulatory mechanism for signal transduction pathways. Recently, an
acylhydrolase previously identified as lysophospholipase has been shown to
function as an acyl protein thioesterase, which catalyzes depalmitoylation of
Galpha proteins as well as Ras. Its amino acid sequence suggested that the
protein is evolutionarily related to neutral lipases and other thioesterases,
but direct structural information was not available. RESULTS: We have solved the
crystal structure of the human putative Galpha-regulatory protein acyl
thioesterase (hAPT1) with a single data set collected from a crystal containing
the wild-type protein. The phases were calculated to 1.8 A resolution based on
anomalous scattering from Br(-) ions introduced in the cryoprotectant solution
in which the crystal was soaked for 20 s. The model was refined against data
extending to a resolution of 1.5 A to an R factor of 18.6%. The enzyme is a
member of the ubiquitous alpha/beta hydrolase family, which includes other
acylhydrolases such as the palmitoyl protein thioesterase (PPT1). CONCLUSIONS:
The human APT1 is closely related to a previously described carboxylesterase
from Pseudomonas fluorescens. The active site contains a catalytic triad of
Ser-114, His-203, and Asp-169. Like carboxylesterase, hAPT1 appears to be
dimeric, although the mutual disposition of molecules in the two dimers differs.
Unlike carboxylesterase, the substrate binding pocket and the active site of
hAPT1 are occluded by the dimer interface, suggesting that the enzyme must
dissociate upon interaction with substrate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 3.
Figure 3. Comparison of the Structures of hAPT1 and CPf(a)
Ribbon presentation of the superposition of Ca atoms of carboxyl
esterase (blue) onto the structure of hAPT1 (gold). Labeled
active site residues are shown by ball and stick models.(b)
Difference distance plot of the corresponding Ca atoms

|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2000,
8,
1137-1146)
copyright 2000.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.S.Pannu,
W.J.Waterreus,
P.Skubák,
I.Sikharulidze,
J.P.Abrahams,
and
R.A.de Graaff
(2011).
Recent advances in the CRANK software suite for experimental phasing.
|
| |
Acta Crystallogr D Biol Crystallogr,
67,
331-337.
|
 |
|
|
|
|
 |
R.J.Read,
and
A.J.McCoy
(2011).
Using SAD data in Phaser.
|
| |
Acta Crystallogr D Biol Crystallogr,
67,
338-344.
|
 |
|
|
|
|
 |
F.J.Dekker,
O.Rocks,
N.Vartak,
S.Menninger,
C.Hedberg,
R.Balamurugan,
S.Wetzel,
S.Renner,
M.Gerauer,
B.Schölermann,
M.Rusch,
J.W.Kramer,
D.Rauh,
G.W.Coates,
L.Brunsveld,
P.I.Bastiaens,
and
H.Waldmann
(2010).
Small-molecule inhibition of APT1 affects Ras localization and signaling.
|
| |
Nat Chem Biol,
6,
449-456.
|
 |
|
|
|
|
 |
P.Skubák,
W.J.Waterreus,
and
N.S.Pannu
(2010).
Multivariate phase combination improves automated crystallographic model building.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
783-788.
|
 |
|
|
|
|
 |
S.K.Parker,
R.M.Barkley,
J.G.Rino,
and
M.L.Vasil
(2009).
Mycobacterium tuberculosis Rv3802c encodes a phospholipase/thioesterase and is inhibited by the antimycobacterial agent tetrahydrolipstatin.
|
| |
PLoS ONE,
4,
e4281.
|
 |
|
|
|
|
 |
C.Dumas,
and
A.van der Lee
(2008).
Macromolecular structure solution by charge flipping.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
864-873.
|
 |
|
|
|
|
 |
A.Lyly,
C.von Schantz,
T.Salonen,
O.Kopra,
J.Saarela,
M.Jauhiainen,
A.Kyttälä,
and
A.Jalanko
(2007).
Glycosylation, transport, and complex formation of palmitoyl protein thioesterase 1 (PPT1)--distinct characteristics in neurons.
|
| |
BMC Cell Biol,
8,
22.
|
 |
|
|
|
|
 |
H.K.Saini,
and
D.Fischer
(2007).
Structural and functional insights into Mimivirus ORFans.
|
| |
BMC Genomics,
8,
115.
|
 |
|
|
|
|
 |
R.Flaumenhaft,
N.Rozenvayn,
D.Feng,
and
A.M.Dvorak
(2007).
SNAP-23 and syntaxin-2 localize to the extracellular surface of the platelet plasma membrane.
|
| |
Blood,
110,
1492-1501.
|
 |
|
|
|
|
 |
S.C.Vose,
N.T.Holland,
B.Eskenazi,
and
J.E.Casida
(2007).
Lysophosphatidylcholine hydrolases of human erythrocytes, lymphocytes, and brain: sensitive targets of conserved specificity for organophosphorus delayed neurotoxicants.
|
| |
Toxicol Appl Pharmacol,
224,
98.
|
 |
|
|
|
|
 |
B.M.Harvey,
H.Hong,
M.A.Jones,
Z.A.Hughes-Thomas,
R.M.Goss,
M.L.Heathcote,
V.M.Bolanos-Garcia,
W.Kroutil,
J.Staunton,
P.F.Leadlay,
and
J.B.Spencer
(2006).
Evidence that a novel thioesterase is responsible for polyketide chain release during biosynthesis of the polyether ionophore monensin.
|
| |
Chembiochem,
7,
1435-1442.
|
 |
|
|
|
|
 |
G.Schneider,
G.Neuberger,
M.Wildpaner,
S.Tian,
I.Berezovsky,
and
F.Eisenhaber
(2006).
Application of a sensitive collection heuristic for very large protein families: evolutionary relationship between adipose triglyceride lipase (ATGL) and classic mammalian lipases.
|
| |
BMC Bioinformatics,
7,
164.
|
 |
|
|
|
|
 |
J.W.Giraldes,
D.L.Akey,
J.D.Kittendorf,
D.H.Sherman,
J.L.Smith,
and
R.A.Fecik
(2006).
Structural and mechanistic insights into polyketide macrolactonization from polyketide-based affinity labels.
|
| |
Nat Chem Biol,
2,
531-536.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Bencharit,
C.C.Edwards,
C.L.Morton,
E.L.Howard-Williams,
P.Kuhn,
P.M.Potter,
and
M.R.Redinbo
(2006).
Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1.
|
| |
J Mol Biol,
363,
201-214.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Pesaresi,
G.Devescovi,
D.Lamba,
V.Venturi,
and
G.Degrassi
(2005).
Isolation, characterization, and heterologous expression of a carboxylesterase of Pseudomonas aeruginosa PAO1.
|
| |
Curr Microbiol,
50,
102-109.
|
 |
|
|
|
|
 |
P.Skubák,
S.Ness,
and
N.S.Pannu
(2005).
Extending the resolution and phase-quality limits in automated model building with iterative refinement.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1626-1635.
|
 |
|
|
|
|
 |
R.Caliandro,
B.Carrozzini,
G.L.Cascarano,
L.De Caro,
C.Giacovazzo,
and
D.Siliqi
(2005).
Phasing at resolution higher than the experimental resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
556-565.
|
 |
|
|
|
|
 |
B.Chakravarty,
Z.Gu,
S.S.Chirala,
S.J.Wakil,
and
F.A.Quiocho
(2004).
Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain.
|
| |
Proc Natl Acad Sci U S A,
101,
15567-15572.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Janda,
Y.Devedjiev,
D.Cooper,
M.Chruszcz,
U.Derewenda,
A.Gabrys,
W.Minor,
A.Joachimiak,
and
Z.S.Derewenda
(2004).
Harvesting the high-hanging fruit: the structure of the YdeN gene product from Bacillus subtilis at 1.8 angstroms resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1101-1107.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.W.Wang,
J.R.Chen,
Y.X.Gu,
C.D.Zheng,
F.Jiang,
and
H.F.Fan
(2004).
Optimizing the error term in direct-method SAD phasing.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1987-1990.
|
 |
|
|
|
|
 |
G.Calero,
P.Gupta,
M.C.Nonato,
S.Tandel,
E.R.Biehl,
S.L.Hofmann,
and
J.Clardy
(2003).
The crystal structure of palmitoyl protein thioesterase-2 (PPT2) reveals the basis for divergent substrate specificities of the two lysosomal thioesterases, PPT1 and PPT2.
|
| |
J Biol Chem,
278,
37957-37964.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Usón,
B.Schmidt,
R.von Bülow,
S.Grimme,
K.von Figura,
M.Dauter,
K.R.Rajashankar,
Z.Dauter,
and
G.M.Sheldrick
(2003).
Locating the anomalous scatterer substructures in halide and sulfur phasing.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
57-66.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Qanbar,
and
M.Bouvier
(2003).
Role of palmitoylation/depalmitoylation reactions in G-protein-coupled receptor function.
|
| |
Pharmacol Ther,
97,
1.
|
 |
|
|
|
|
 |
E.Girard,
L.Chantalat,
J.Vicat,
and
R.Kahn
(2002).
Gd-HPDO3A, a complex to obtain high-phasing-power heavy-atom derivatives for SAD and MAD experiments: results with tetragonal hen egg-white lysozyme.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
1-9.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.D.Bruner,
T.Weber,
R.M.Kohli,
D.Schwarzer,
M.A.Marahiel,
C.T.Walsh,
and
M.T.Stubbs
(2002).
Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE.
|
| |
Structure,
10,
301-310.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Dauter,
M.Dauter,
and
E.Dodson
(2002).
Jolly SAD.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
494-506.
|
 |
|
|
|
|
 |
Z.Dauter
(2002).
One-and-a-half wavelength approach.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
1958-1967.
|
 |
|
|
|
|
 |
S.C.Tsai,
L.J.Miercke,
J.Krucinski,
R.Gokhale,
J.C.Chen,
P.G.Foster,
D.E.Cane,
C.Khosla,
and
R.M.Stroud
(2001).
Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
|
| |
Proc Natl Acad Sci U S A,
98,
14808-14813.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Dauter,
and
M.Dauter
(2001).
Entering a new phase: using solvent halide ions in protein structure determination.
|
| |
Structure,
9,
R21-R26.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

