 |
PDBsum entry 1fb5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.3.3
- ornithine carbamoyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Urea Cycle and Arginine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
carbamoyl phosphate + L-ornithine = L-citrulline + phosphate + H+
|
 |
 |
 |
 |
 |
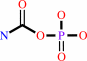 carbamoyl phosphate
carbamoyl phosphate
|
+
|
L-ornithine
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
=
|
 L-citrulline
L-citrulline
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Org Biomol Chem
1:3178-3185
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Functional and structural characterization of ovine ornithine transcarbamoylase.
|
|
A.De Gregorio,
R.Battistutta,
N.Arena,
M.Panzalorto,
P.Francescato,
G.Valentini,
G.Bruno,
G.Zanotti.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ornithine transcarbamoylase from ovine liver has been purified to homogeneity.
Like all anabolic OTCs, the ovine enzyme is a trimer, constituted by identical
subunits of 34 kDa. Sequence analysis of the 54 N-terminal residues of ovine OTC
shows a high degree of homology with the human enzyme. The optimum pH and the
Michaelis constants for the catalytic reaction were determined. The ovine enzyme
is the most thermostable one among mammals OTCs, its critical temperature being
6 degrees C higher than those measured for the other enzymes. The enzyme has
been crystallised and the structure determined at 3.5 A resolution. Crystals
belong to the cubic P4(3)32 space group, with a = b = c = 184.7 A and a solvent
content of about 80%. There is no evidence of any ligand in the active site
cavity, indicating that the crystals contain an unliganded or T state of the
enzyme. The unliganded OTCase enzyme adopts a trimeric structure which, in the
crystal, presents a three-fold axis coincident with the crystallographic one.
The conformation of each monomer in the trimer is quite similar to that of the
liganded human protein, with the exception of a few loops, directly interacting
with the substrate(s), which are able to induce a rearrangement of the
quaternary organisation of the trimer, that accounts for the cooperative
behaviour of the enzyme following the binding of the substrates.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.Moradian,
C.Garen,
L.Cherney,
M.Cherney,
and
M.N.James
(2006).
Expression, purification, crystallization and preliminary X-ray analysis of two arginine-biosynthetic enzymes from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
986-988.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

