 |
PDBsum entry 1f11

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Immune system
|
PDB id
|
|

|

|
1f11
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Immune system
|
 |
|
Title:
|
 |
F124 fab fragment from a monoclonal anti-pres2 antibody
|
|
Structure:
|
 |
F124 immunoglobulin (kappa light chain). Chain: a, c. Fragment: fab fragment (unp p01665 residues 1-111, p01837 residues 1- 106). F124 immunoglobulin (igg1 heavy chain). Chain: b, d. Fragment: fab fragment (unp q65zr6 residues 18-134, p01869 residues 2-102)
|
|
Source:
|
 |
Mus musculus. Mouse. Organism_taxid: 10090. Organism_taxid: 10090
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
3.00Å
|
R-factor:
|
0.184
|
R-free:
|
0.271
|
|
|
Authors:
|
 |
F.A.Saul,B.Vulliez-Le Normand,M.Passafiume,M.M.Riottot,G.A.Bentley
|
Key ref:

|
 |
F.A.Saul
et al.
(2000).
Structure of the Fab fragment from F124, a monoclonal antibody specific for hepatitis B surface antigen.
Acta Crystallogr D Biol Crystallogr,
56,
945-951.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
18-May-00
|
Release date:
|
30-Aug-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P01665
(KV3AD_MOUSE) -
Ig kappa chain V-III region PC 7043 from Mus musculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
111 a.a.
216 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
56:945-951
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of the Fab fragment from F124, a monoclonal antibody specific for hepatitis B surface antigen.
|
|
F.A.Saul,
B.Vulliez-Le Normand,
M.Passafiume,
M.M.Riottot,
G.A.Bentley.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the Fab fragment from the monoclonal anti-preS2
antibody F124 (IgG1,kappa) has been solved by molecular replacement and refined
at 3.0 A resolution. The Fab crystallizes with two independent molecules in the
asymmetric unit. F124 recognizes an epitope contained within the preS2 segment
between residues 120 and 132 of the surface antigen of hepatitis B virus. The
antibody shows a high affinity for the glycan N-linked to Asn123, but it also
cross-reacts with the non-glycosylated peptide fragment 120-132. Although
crystallization was performed in the presence of an eightfold excess of the
cross-reactive peptide, no evidence for the ligand was found in the
antigen-binding site, which is close to a neighbouring molecule in the crystal
lattice. The antigen-binding site has a groove-like topology which is modulated
with pocket-like cavities. It is characterized by a large number of tyrosine and
aspartate residues. The importance of germ-line mutations at the binding site is
discussed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Ramachandran plot for the two independent molecules of
Fab F124. A total of 83.6% of the non-glycine residues fall in
the most favoured regions and 15.6% in the additional allowed
region. AlaL51, which is in an unfavoured region, has dihedral
angles that are characteristic of a 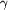 -turn
(Milner-White et al., 1988[Milner-White, E. J., Ross, B. M.,
Ismail, R., Belhady-Mostefa, K. & Poet, R. (1988). J. Mol. Biol.
204, 777-782.]). Residues in the unfavoured regions are
labelled; glycines are shown as triangles. (Produced by the
program PROCHECK; Laskowski et al., 1993 [Laskowski, R. A.,
McArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl.
Cryst. -turn
(Milner-White et al., 1988[Milner-White, E. J., Ross, B. M.,
Ismail, R., Belhady-Mostefa, K. & Poet, R. (1988). J. Mol. Biol.
204, 777-782.]). Residues in the unfavoured regions are
labelled; glycines are shown as triangles. (Produced by the
program PROCHECK; Laskowski et al., 1993 [Laskowski, R. A.,
McArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl.
Cryst.
|
 |
Figure 5.
Figure 5 A stereoview of the antigen-binding site colour-coded
for electrostatic potential; red represents positive and blue
negative. (Prepared with the program GRASP; Nicholls et al.,
1991[Nicholls, A., Sharp, K. A. & Honig, B. (1991). Proteins
Struct. Funct. Genet. 11, 281-296.].)
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2000,
56,
945-951)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.L.Longenecker,
Q.Ruan,
E.H.Fry,
S.C.Saldana,
S.E.Brophy,
P.L.Richardson,
and
S.Y.Tetin
(2009).
Crystal structure and thermodynamic analysis of diagnostic mAb 106.3 complexed with BNP 5-13 (C10A).
|
| |
Proteins,
76,
536-547.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.R.Watts,
G.Cardone,
J.G.Vethanayagam,
N.Cheng,
C.Hultgren,
S.J.Stahl,
A.C.Steven,
M.Sällberg,
and
P.T.Wingfield
(2008).
Non-canonical binding of an antibody resembling a naïve B cell receptor immunoglobulin to hepatitis B virus capsids.
|
| |
J Mol Biol,
379,
1119-1129.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
| |

