 |
PDBsum entry 1eox

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Carbohydrate metabolism
|
PDB id
|
|

|

|
1eox
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.37
- sedoheptulose-bisphosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Calvin Cycle
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-sedoheptulose 1,7-bisphosphate + H2O = D-sedoheptulose 7-phosphate + phosphate
|
 |
 |
 |
 |
 |
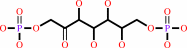 D-sedoheptulose 1,7-bisphosphate
D-sedoheptulose 1,7-bisphosphate
|
+
|
H2O
|
=
|
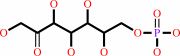 D-sedoheptulose 7-phosphate
D-sedoheptulose 7-phosphate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Plant J
10:553-560
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Identification of a potential redox-sensitive interdomain disulfide in the sedoheptulose bisphosphatase of Chlamydomonas reinhardtii.
|
|
L.E.Anderson,
H.C.Huppe,
A.D.Li,
F.J.Stevens.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In the stimulated three-dimensional structure of the Chlamydomonas reinhardtii
sedoheptulose bisphosphatase (EC 3.1.3.37) there are two cysteine residues close
enough to one another to form a redox-sensitive disulfide bond which would
cross-link the nucleotide and carbon substrate domains. Examination of the redox
modulation of this sedoheptulose bisphosphatase confirms that it resembles the
higher plant enzyme in being activated by reduction. In the wheat and
Arabidopsis enzymes, for which there is sequence information and which, like the
Chlamydomonas enzyme, can be modeled, both redox-sensitive Cys residues appear
to be located on the regulatory nucleotide-binding domain. Apparently different
Cys residues are involved in modulation in the algal and higher plant
sedoheptulose bisphosphatases.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Kozaki,
K.Mayumi,
and
Y.Sasaki
(2001).
Thiol-disulfide exchange between nuclear-encoded and chloroplast-encoded subunits of pea acetyl-CoA carboxylase.
|
| |
J Biol Chem,
276,
39919-39925.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

