
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.6.1.1.20
- phenylalanine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Phe) + L-phenylalanine + ATP = L-phenylalanyl-tRNA(Phe) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
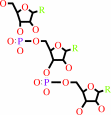 tRNA(Phe)
tRNA(Phe)
|
+
|
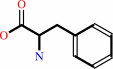 L-phenylalanine
L-phenylalanine
|
+
|
 ATP
ATP
|
=
|
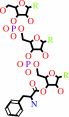 L-phenylalanyl-tRNA(Phe)
L-phenylalanyl-tRNA(Phe)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
5:59-68
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe.
|
|
Y.Goldgur,
L.Mosyak,
L.Reshetnikova,
V.Ankilova,
O.Lavrik,
S.Khodyreva,
M.Safro.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: In the translation of the genetic code each aminoacyl-tRNA
synthetase (aaRS) must recognize its own (cognate) tRNA and attach the
corresponding amino acid to the acceptor end of tRNA, discriminating all the
others. The(alphabeta)2 phenylalanyl-tRNA synthetase (PheRS) is one of the most
complex enzymes in the aaRS family and is characterized by anomalous charging
properties. Structurally, the enzyme belongs to class II aaRSs, as its catalytic
domain is built around an antiparallel beta sheet, but functionally it resembles
class I as it aminoacylates the 2'OH of the terminal ribose of tRNA (class II
aaRSs aminoacylate the 3'OH). With the availability of the three-dimensional
structure of the complex between multisubunit PheRS and tRNAPhe, a fuller
picture of the specific tRNA-aaRS interactions is beginning to emerge. RESULTS:
The crystal structure of Thermus thermophilus PheRS complexed with cognate tRNA
has been solved at 3.28 A resolution. It reveals that one tRNAPhe molecule binds
across all four PheRS subunits. The interactions of PheRS with tRNA stabilize
the flexible N-terminal part of the alpha subunit, which appeared to form the
enzyme's 11th domain, comprising a coiled-coil structure (helical arm) built up
of two long antiparallel alpha helices. The helical arms are similar to those
observed in SerRS and are in the same relative orientation with respect to the
catalytic domain. Anticodon recognition upon tRNA binding is performed by the B8
domain, the structure of which is similar to that of the RNA-binding domain
(RBD) of the small spliceosomal protein U1A. The Th. thermophilus PheRS
approaches the anticodon loop from the minor groove side. CONCLUSIONS: The mode
of interactions with tRNA explains the absolute necessity for the (alphabeta)2
architecture of PheRS. The interactions of tRNAPhe with PheRS and particularly
with the coiled-coil domain of the alpha subunit result in conformational
changes in TPsiC and D loops seen by comparison with uncomplexed yeast tRNAPhe.
The tRNAPhe is a newly recognized type of RNA molecule specifically interacting
with the RBD fold. In addition, a new type of anticodon-binding domain emerges
in the aaRS family. The uniqueness of PheRS in charging 2'OH of tRNA is dictated
by the size of its adenine-binding pocket and by the local conformation of the
tRNA's CCA end.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 4.
Figure 4. View of the CCA end of tRNA^Phe in the
active-site cavity of the enzyme. The dashed lines represent the
hydrogen bonds. The letters A and B before residue numbers
indicate the enzyme subunits. The relatively short distance (
 vert,
similar 3.8 Å) between the N6 group of A73 (ADE73) and the
phosphate group of C72 (CYT72) indicates that their
intramolecular interaction may help to stabilize the
conformation of the CCA end in a way that resembles tRNA^Gln
[4]. However, the conformation of tRNA^Phe in this region
differs from that of tRNA^Gln. The letters A and B before
residue numbers indicate the enzyme subunits. (Figure was drawn
using MOLSCRIPT [36].) vert,
similar 3.8 Å) between the N6 group of A73 (ADE73) and the
phosphate group of C72 (CYT72) indicates that their
intramolecular interaction may help to stabilize the
conformation of the CCA end in a way that resembles tRNA^Gln
[4]. However, the conformation of tRNA^Phe in this region
differs from that of tRNA^Gln. The letters A and B before
residue numbers indicate the enzyme subunits. (Figure was drawn
using MOLSCRIPT [36].)
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1997,
5,
59-68)
copyright 1997.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Mermershtain,
I.Finarov,
L.Klipcan,
N.Kessler,
H.Rozenberg,
and
M.G.Safro
(2011).
Idiosyncrasy and identity in the prokaryotic phe-system: crystal structure of E. coli phenylalanyl-tRNA synthetase complexed with phenylalanine and AMP.
|
| |
Protein Sci,
20,
160-167.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Finarov,
N.Moor,
N.Kessler,
L.Klipcan,
and
M.G.Safro
(2010).
Structure of human cytosolic phenylalanyl-tRNA synthetase: evidence for kingdom-specific design of the active sites and tRNA binding patterns.
|
| |
Structure,
18,
343-353.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Shaul,
D.Berel,
Y.Benjamini,
and
D.Graur
(2010).
Revisiting the operational RNA code for amino acids: Ensemble attributes and their implications.
|
| |
RNA,
16,
141-153.
|
 |
|
|
|
|
 |
I.A.Vasil'eva,
E.A.Semenova,
and
N.A.Moor
(2009).
Interaction of human phenylalanyl-tRNA synthetase with specific tRNA according to thiophosphate footprinting.
|
| |
Biochemistry (Mosc),
74,
175-185.
|
 |
|
|
|
|
 |
I.Finarov,
N.Moor,
N.Kessler,
and
M.Safro
(2009).
Crystallization and X-ray analysis of human cytoplasmic phenylalanyl-tRNA synthetase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
93-97.
|
 |
|
|
|
|
 |
J.Ling,
B.R.So,
S.S.Yadavalli,
H.Roy,
S.Shoji,
K.Fredrick,
K.Musier-Forsyth,
and
M.Ibba
(2009).
Resampling and editing of mischarged tRNA prior to translation elongation.
|
| |
Mol Cell,
33,
654-660.
|
 |
|
|
|
|
 |
K.Nozawa,
P.O'Donoghue,
S.Gundllapalli,
Y.Araiso,
R.Ishitani,
T.Umehara,
D.Söll,
and
O.Nureki
(2009).
Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality.
|
| |
Nature,
457,
1163-1167.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Goto-Ito,
T.Ito,
M.Kuratani,
Y.Bessho,
and
S.Yokoyama
(2009).
Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation.
|
| |
Nat Struct Mol Biol,
16,
1109-1115.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Klipcan,
I.Levin,
N.Kessler,
N.Moor,
I.Finarov,
and
M.Safro
(2008).
The tRNA-induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase.
|
| |
Structure,
16,
1095-1104.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.F.Egea,
J.Napetschnig,
P.Walter,
and
R.M.Stroud
(2008).
Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
|
| |
PLoS ONE,
3,
e3528.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.An,
G.Barany,
and
K.Musier-Forsyth
(2008).
Evolution of acceptor stem tRNA recognition by class II prolyl-tRNA synthetase.
|
| |
Nucleic Acids Res,
36,
2514-2521.
|
 |
|
|
|
|
 |
S.I.Hauenstein,
Y.M.Hou,
and
J.J.Perona
(2008).
The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity.
|
| |
J Biol Chem,
283,
21997-22006.
|
 |
|
|
|
|
 |
S.N.Rodin,
and
A.S.Rodin
(2008).
On the origin of the genetic code: signatures of its primordial complementarity in tRNAs and aminoacyl-tRNA synthetases.
|
| |
Heredity,
100,
341-355.
|
 |
|
|
|
|
 |
A.Raymond,
and
S.Shuman
(2007).
Deinococcus radiodurans RNA ligase exemplifies a novel ligase clade with a distinctive N-terminal module that is important for 5'-PO4 nick sealing and ligase adenylylation but dispensable for phosphodiester formation at an adenylylated nick.
|
| |
Nucleic Acids Res,
35,
839-849.
|
 |
|
|
|
|
 |
I.A.Vasil'eva,
and
N.A.Moor
(2007).
Interaction of aminoacyl-tRNA synthetases with tRNA: general principles and distinguishing characteristics of the high-molecular-weight substrate recognition.
|
| |
Biochemistry (Mosc),
72,
247-263.
|
 |
|
|
|
|
 |
I.Levin,
N.Kessler,
N.Moor,
L.Klipcan,
E.Koc,
P.Templeton,
L.Spremulli,
and
M.Safro
(2007).
Purification, crystallization and preliminary X-ray characterization of a human mitochondrial phenylalanyl-tRNA synthetase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
761-764.
|
 |
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2007).
Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea.
|
| |
Nat Struct Mol Biol,
14,
272-279.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Tyagi,
and
D.H.Mathews
(2007).
Predicting helical coaxial stacking in RNA multibranch loops.
|
| |
RNA,
13,
939-951.
|
 |
|
|
|
|
 |
C.Hoang,
J.Chen,
C.A.Vizthum,
J.M.Kandel,
C.S.Hamilton,
E.G.Mueller,
and
A.R.Ferré-D'Amaré
(2006).
Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure.
|
| |
Mol Cell,
24,
535-545.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Kodama,
S.Fukuzawa,
K.Sakamoto,
H.Nakayama,
T.Kigawa,
T.Yabuki,
N.Matsuda,
M.Shirouzu,
K.Takio,
K.Tachibana,
and
S.Yokoyama
(2006).
A new protein engineering approach combining chemistry and biology, part I; site-specific incorporation of 4-iodo-L-phenylalanine in vitro by using misacylated suppressor tRNAPhe.
|
| |
Chembiochem,
7,
1577-1581.
|
 |
|
|
|
|
 |
S.N.Rodin,
and
A.S.Rodin
(2006).
Partitioning of aminoacyl-tRNA synthetases in two classes could have been encoded in a strand-symmetric RNA world.
|
| |
DNA Cell Biol,
25,
617-626.
|
 |
|
|
|
|
 |
S.Wang,
Y.Hu,
M.T.Overgaard,
F.V.Karginov,
O.C.Uhlenbeck,
and
D.B.McKay
(2006).
The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold.
|
| |
RNA,
12,
959-967.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Meka,
F.Werner,
S.C.Cordell,
S.Onesti,
and
P.Brick
(2005).
Crystal structure and RNA binding of the Rpb4/Rpb7 subunits of human RNA polymerase II.
|
| |
Nucleic Acids Res,
33,
6435-6444.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Sakurai,
T.Ohtsuki,
and
K.Watanabe
(2005).
Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm.
|
| |
Nucleic Acids Res,
33,
1653-1661.
|
 |
|
|
|
|
 |
O.Kotik-Kogan,
N.Moor,
D.Tworowski,
and
M.Safro
(2005).
Structural basis for discrimination of L-phenylalanine from L-tyrosine by phenylalanyl-tRNA synthetase.
|
| |
Structure,
13,
1799-1807.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Petry,
D.E.Brodersen,
F.V.Murphy,
C.M.Dunham,
M.Selmer,
M.J.Tarry,
A.C.Kelley,
and
V.Ramakrishnan
(2005).
Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon.
|
| |
Cell,
123,
1255-1266.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Zhang,
L.Wang,
P.G.Schultz,
and
I.A.Wilson
(2005).
Crystal structures of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine.
|
| |
Protein Sci,
14,
1340-1349.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Martins,
and
S.Shuman
(2004).
An RNA ligase from Deinococcus radiodurans.
|
| |
J Biol Chem,
279,
50654-50661.
|
 |
|
|
|
|
 |
D.Beyer,
H.P.Kroll,
R.Endermann,
G.Schiffer,
S.Siegel,
M.Bauser,
J.Pohlmann,
M.Brands,
K.Ziegelbauer,
D.Haebich,
C.Eymann,
and
H.Brötz-Oesterhelt
(2004).
New class of bacterial phenylalanyl-tRNA synthetase inhibitors with high potency and broad-spectrum activity.
|
| |
Antimicrob Agents Chemother,
48,
525-532.
|
 |
|
|
|
|
 |
H.Roy,
J.Ling,
M.Irnov,
and
M.Ibba
(2004).
Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase.
|
| |
EMBO J,
23,
4639-4648.
|
 |
|
|
|
|
 |
D.L.Theobald,
R.M.Mitton-Fry,
and
D.S.Wuttke
(2003).
Nucleic acid recognition by OB-fold proteins.
|
| |
Annu Rev Biophys Biomol Struct,
32,
115-133.
|
 |
|
|
|
|
 |
D.Tworowski,
and
M.Safro
(2003).
The long-range electrostatic interactions control tRNA-aminoacyl-tRNA synthetase complex formation.
|
| |
Protein Sci,
12,
1247-1251.
|
 |
|
|
|
|
 |
M.Francin,
and
M.Mirande
(2003).
Functional dissection of the eukaryotic-specific tRNA-interacting factor of lysyl-tRNA synthetase.
|
| |
J Biol Chem,
278,
1472-1479.
|
 |
|
|
|
|
 |
W.Xie,
X.Liu,
and
R.H.Huang
(2003).
Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate.
|
| |
Nat Struct Biol,
10,
781-788.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.A.Vasil'eva,
V.N.Ankilova,
O.I.Lavrik,
and
N.A.Moor
(2002).
tRNA discrimination by T. thermophilus phenylalanyl-tRNA synthetase at the binding step.
|
| |
J Mol Recognit,
15,
188-196.
|
 |
|
|
|
|
 |
P.S.Klosterman,
M.Tamura,
S.R.Holbrook,
and
S.E.Brenner
(2002).
SCOR: a Structural Classification of RNA database.
|
| |
Nucleic Acids Res,
30,
392-394.
|
 |
|
|
|
|
 |
C.Hoang,
and
A.R.Ferré-D'Amaré
(2001).
Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme.
|
| |
Cell,
107,
929-939.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Todone,
P.Brick,
F.Werner,
R.O.Weinzierl,
and
S.Onesti
(2001).
Structure of an archaeal homolog of the eukaryotic RNA polymerase II RPB4/RPB7 complex.
|
| |
Mol Cell,
8,
1137-1143.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Renault,
P.Kerjan,
S.Pasqualato,
J.Ménétrey,
J.C.Robinson,
S.Kawaguchi,
D.G.Vassylyev,
S.Yokoyama,
M.Mirande,
and
J.Cherfils
(2001).
Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry.
|
| |
EMBO J,
20,
570-578.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Ribas de Pouplana,
and
P.Schimmel
(2001).
Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem.
|
| |
Cell,
104,
191-193.
|
 |
|
|
|
|
 |
L.Ribas de Pouplana,
and
P.Schimmel
(2001).
Aminoacyl-tRNA synthetases: potential markers of genetic code development.
|
| |
Trends Biochem Sci,
26,
591-596.
|
 |
|
|
|
|
 |
N.A.Moor,
V.N.Ankilova,
O.I.Lavrik,
and
A.Favre
(2001).
Determination of tRNA(Phe) nucleotides contacting the subunits of Thermus thermophilus phenylalanyl-tRNA synthetase by photoaffinity crosslinking.
|
| |
Biochim Biophys Acta,
1518,
226-236.
|
 |
|
|
|
|
 |
S.Raveh,
J.Vinh,
J.Rossier,
F.Agou,
and
M.Véron
(2001).
Peptidic determinants and structural model of human NDP kinase B (Nm23-H2) bound to single-stranded DNA.
|
| |
Biochemistry,
40,
5882-5893.
|
 |
|
|
|
|
 |
T.Inoue,
K.Kizawa,
and
M.Ito
(2001).
Characterization of soluble protein extracts from keratinized tissues: identification of ubiquitin universally distributed in hair, nail, and stratum corneum.
|
| |
Biosci Biotechnol Biochem,
65,
895-900.
|
 |
|
|
|
|
 |
T.L.Hendrickson
(2001).
Recognizing the D-loop of transfer RNAs.
|
| |
Proc Natl Acad Sci U S A,
98,
13473-13475.
|
 |
|
|
|
|
 |
A.A.Antson
(2000).
Single-stranded-RNA binding proteins.
|
| |
Curr Opin Struct Biol,
10,
87-94.
|
 |
|
|
|
|
 |
A.D.Frankel
(2000).
Fitting peptides into the RNA world.
|
| |
Curr Opin Struct Biol,
10,
332-340.
|
 |
|
|
|
|
 |
B.Burke,
F.Yang,
F.Chen,
C.Stehlin,
B.Chan,
and
K.Musier-Forsyth
(2000).
Evolutionary coadaptation of the motif 2--acceptor stem interaction in the class II prolyl-tRNA synthetase system.
|
| |
Biochemistry,
39,
15540-15547.
|
 |
|
|
|
|
 |
G.Martin,
W.Keller,
and
S.Doublié
(2000).
Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP.
|
| |
EMBO J,
19,
4193-4203.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Sugiura,
O.Nureki,
Y.Ugaji-Yoshikawa,
S.Kuwabara,
A.Shimada,
M.Tateno,
B.Lorber,
R.Giegé,
D.Moras,
S.Yokoyama,
and
M.Konno
(2000).
The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.
|
| |
Structure,
8,
197-208.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Pleiss,
A.D.Wolfson,
and
O.C.Uhlenbeck
(2000).
Mapping contacts between Escherichia coli alanyl tRNA synthetase and 2' hydroxyls using a complete tRNA molecule.
|
| |
Biochemistry,
39,
8250-8258.
|
 |
|
|
|
|
 |
M.A.Swairjo,
A.J.Morales,
C.C.Wang,
A.R.Ortiz,
and
P.Schimmel
(2000).
Crystal structure of trbp111: a structure-specific tRNA-binding protein.
|
| |
EMBO J,
19,
6287-6298.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Ibba,
and
D.Soll
(2000).
Aminoacyl-tRNA synthesis.
|
| |
Annu Rev Biochem,
69,
617-650.
|
 |
|
|
|
|
 |
M.Kaminska,
M.Deniziak,
P.Kerjan,
J.Barciszewski,
and
M.Mirande
(2000).
A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation.
|
| |
EMBO J,
19,
6908-6917.
|
 |
|
|
|
|
 |
T.A.Nissan,
and
J.J.Perona
(2000).
Alternative designs for construction of the class II transfer RNA tertiary core.
|
| |
RNA,
6,
1585-1596.
|
 |
|
|
|
|
 |
V.Guez,
S.Nair,
A.Chaffotte,
and
H.Bedouelle
(2000).
The anticodon-binding domain of tyrosyl-tRNA synthetase: state of folding and origin of the crystallographic disorder.
|
| |
Biochemistry,
39,
1739-1747.
|
 |
|
|
|
|
 |
J.C.Nix,
A.R.Newhoff,
and
C.Wilson
(1999).
Preliminary crystallographic characterization of an in vitro evolved biotin-binding RNA pseudoknot.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
323-325.
|
 |
|
|
|
|
 |
L.Jermutus,
V.Guez,
and
H.Bedouelle
(1999).
Disordered C-terminal domain of tyrosyl-tRNA synthetase: secondary structure prediction.
|
| |
Biochimie,
81,
235-244.
|
 |
|
|
|
|
 |
P.J.Beuning,
and
K.Musier-Forsyth
(1999).
Transfer RNA recognition by aminoacyl-tRNA synthetases.
|
| |
Biopolymers,
52,
1.
|
 |
|
|
|
|
 |
T.A.Nissan,
B.Oliphant,
and
J.J.Perona
(1999).
An engineered class I transfer RNA with a class II tertiary fold.
|
| |
RNA,
5,
434-445.
|
 |
|
|
|
|
 |
Y.Zhao,
D.Jeruzalmi,
I.Moarefi,
L.Leighton,
R.Lasken,
and
J.Kuriyan
(1999).
Crystal structure of an archaebacterial DNA polymerase.
|
| |
Structure,
7,
1189-1199.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.D.Frankel,
and
C.A.Smith
(1998).
Induced folding in RNA-protein recognition: more than a simple molecular handshake.
|
| |
Cell,
92,
149-151.
|
 |
|
|
|
|
 |
G.Simos,
A.Sauer,
F.Fasiolo,
and
E.C.Hurt
(1998).
A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases.
|
| |
Mol Cell,
1,
235-242.
|
 |
|
|
|
|
 |
M.Frugier,
M.Helm,
B.Felden,
R.Giegé,
and
C.Florentz
(1998).
Sequences outside recognition sets are not neutral for tRNA aminoacylation. Evidence for nonpermissive combinations of nucleotides in the acceptor stem of yeast tRNAPhe.
|
| |
J Biol Chem,
273,
11605-11610.
|
 |
|
|
|
|
 |
W.H.McClain,
J.Schneider,
S.Bhattacharya,
and
K.Gabriel
(1998).
The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation.
|
| |
Proc Natl Acad Sci U S A,
95,
460-465.
|
 |
|
|
|
|
 |
A.Ramos,
and
G.Varani
(1997).
Structure of the acceptor stem of Escherichia coli tRNA Ala: role of the G3.U70 base pair in synthetase recognition.
|
| |
Nucleic Acids Res,
25,
2083-2090.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.L.Hansen,
A.M.Long,
and
S.C.Schultz
(1997).
Structure of the RNA-dependent RNA polymerase of poliovirus.
|
| |
Structure,
5,
1109-1122.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Cusack
(1997).
Aminoacyl-tRNA synthetases.
|
| |
Curr Opin Struct Biol,
7,
881-889.
|
 |
|
|
|
|
 |
S.Quevillon,
F.Agou,
J.C.Robinson,
and
M.Mirande
(1997).
The p43 component of the mammalian multi-synthetase complex is likely to be the precursor of the endothelial monocyte-activating polypeptide II cytokine.
|
| |
J Biol Chem,
272,
32573-32579.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

