 |
PDBsum entry 1e7r

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.271
- GDP-L-fucose synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
GDP-L-Fucose and GDP-mannose Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
GDP-beta-L-fucose + NADP+ = GDP-4-dehydro-alpha-D-rhamnose + NADPH + H+
|
 |
 |
 |
 |
 |
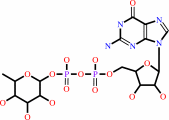 GDP-beta-L-fucose
GDP-beta-L-fucose
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
GDP-4-dehydro-alpha-D-rhamnose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
303:77-91
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Probing the catalytic mechanism of GDP-4-keto-6-deoxy-d-mannose Epimerase/Reductase by kinetic and crystallographic characterization of site-specific mutants.
|
|
C.Rosano,
A.Bisso,
G.Izzo,
M.Tonetti,
L.Sturla,
A.De Flora,
M.Bolognesi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
GDP-4-keto-6-deoxy-d-mannose epimerase/reductase is a bifunctional enzyme
responsible for the last step in the biosynthesis of GDP-l-fucose, the substrate
of fucosyl transferases. Several cell-surface antigens, including the leukocyte
Lewis system and cell-surface antigens in pathogenic bacteria, depend on the
availability of GDP-l-fucose for their expression. Therefore, the enzyme is a
potential target for therapy in pathological states depending on
selectin-mediated cell-to-cell interactions. Previous crystallographic
investigations have shown that GDP-4-keto-6-deoxy-d-mannose epimerase/reductase
belongs to the short-chain dehydrogenase/reductase protein homology family. The
enzyme active-site region is at the interface of an N-terminal NADPH-binding
domain and a C-terminal domain, held to bind the substrate. The design,
expression and functional characterization of seven site-specific mutant forms
of GDP-4-keto-6-deoxy-d-mannose epimerase/reductase are reported here. In
parallel, the crystal structures of the native holoenzyme and of three mutants
(Ser107Ala, Tyr136Glu and Lys140Arg) have been investigated and refined at 1.
45-1.60 A resolution, based on synchrotron data (R-factors range between 12.6 %
and 13.9 %). The refined protein models show that besides the active-site
residues Ser107, Tyr136 and Lys140, whose mutations impair the overall enzymatic
activity and may affect the coenzyme binding mode, side-chains capable of proton
exchange, located around the expected substrate (GDP-4-keto-6-deoxy-d-mannose)
binding pocket, are selectively required during the epimerization and reduction
steps. Among these, Cys109 and His179 may play a primary role in proton exchange
between the enzyme and the epimerization catalytic intermediates. Finally, the
additional role of mutated active-site residues involved in substrate
recognition and in enzyme stability has been analyzed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The GDP- Image -fucose biosynthetic pathway and
the products obtained after NaBH[4] reduction of the
intermediate compounds. GMD, GDP- Image -mannose 4,6
dehydratase; GMER, GDP-4-keto-6-deoxy- Image -mannose
epimerase/reductase; R-, GDP.
|
 |
Figure 6.
Figure 6. Stereo view of the proposed binding mode for a
nucleotide-sugar molecule (shown as a space-filling model)
relative to active-site residues discussed in the text. The C-2
and C-3 centers can be recognized as those closest to His179
side-chain; the C-4 center falls next to the nicotinamide
carboxamido group.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
303,
77-91)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.R.Kinjo,
and
H.Nakamura
(2008).
Nature of protein family signatures: insights from singular value analysis of position-specific scoring matrices.
|
| |
PLoS ONE,
3,
e1963.
|
 |
|
|
|
|
 |
B.Liu,
Y.A.Knirel,
L.Feng,
A.V.Perepelov,
S.N.Senchenkova,
Q.Wang,
P.R.Reeves,
and
L.Wang
(2008).
Structure and genetics of Shigella O antigens.
|
| |
FEMS Microbiol Rev,
32,
627-653.
|
 |
|
|
|
|
 |
C.J.Thibodeaux,
C.E.Melançon,
and
H.W.Liu
(2008).
Natural-product sugar biosynthesis and enzymatic glycodiversification.
|
| |
Angew Chem Int Ed Engl,
47,
9814-9859.
|
 |
|
|
|
|
 |
K.Imada,
T.Tamura,
R.Takenaka,
I.Kobayashi,
K.Namba,
and
K.Inagaki
(2008).
Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity.
|
| |
Proteins,
70,
63-71.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.L.Kavanagh,
H.Jörnvall,
B.Persson,
and
U.Oppermann
(2008).
Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes.
|
| |
Cell Mol Life Sci,
65,
3895-3906.
|
 |
|
|
|
|
 |
C.Louime,
M.Abazinge,
E.Johnson,
L.Latinwo,
C.Ikediobi,
and
A.M.Clark
(2007).
Molecular cloning and biochemical characterization of a family-9 endoglucanase with an unusual structure from the gliding bacteria Cytophaga hutchinsonii.
|
| |
Appl Biochem Biotechnol,
141,
127-138.
|
 |
|
|
|
|
 |
J.D.King,
N.J.Harmer,
A.Preston,
C.M.Palmer,
M.Rejzek,
R.A.Field,
T.L.Blundell,
and
D.J.Maskell
(2007).
Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
|
| |
J Mol Biol,
374,
749-763.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Rhomberg,
C.Fuchsluger,
D.Rendić,
K.Paschinger,
V.Jantsch,
P.Kosma,
and
I.B.Wilson
(2006).
Reconstitution in vitro of the GDP-fucose biosynthetic pathways of Caenorhabditis elegans and Drosophila melanogaster.
|
| |
FEBS J,
273,
2244-2256.
|
 |
|
|
|
|
 |
B.A.Wolucka,
and
M.Van Montagu
(2003).
GDP-mannose 3',5'-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants.
|
| |
J Biol Chem,
278,
47483-47490.
|
 |
|
|
|
|
 |
L.S.Forsberg,
K.D.Noel,
J.Box,
and
R.W.Carlson
(2003).
Genetic locus and structural characterization of the biochemical defect in the O-antigenic polysaccharide of the symbiotically deficient Rhizobium etli mutant, CE166. Replacement of N-acetylquinovosamine with its hexosyl-4-ulose precursor.
|
| |
J Biol Chem,
278,
51347-51359.
|
 |
|
|
|
|
 |
C.A.Bottoms,
P.E.Smith,
and
J.J.Tanner
(2002).
A structurally conserved water molecule in Rossmann dinucleotide-binding domains.
|
| |
Protein Sci,
11,
2125-2137.
|
 |
|
|
|
|
 |
C.Creuzenet,
R.V.Urbanic,
and
J.S.Lam
(2002).
Structure-function studies of two novel UDP-GlcNAc C6 dehydratases/C4 reductases. Variation from the SYK dogma.
|
| |
J Biol Chem,
277,
26769-26778.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

