 |
PDBsum entry 1e4m

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.147
- thioglucosidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a thioglucoside + H2O = a sugar + a thiol
|
 |
 |
 |
 |
 |
thioglucoside
Bound ligand (Het Group name = )
matches with 84.62% similarity
|
+
|
H2O
|
=
|
sugar
|
+
|
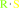 thiol
thiol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
275:39385-39393
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
High resolution X-ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base.
|
|
W.P.Burmeister,
S.Cottaz,
P.Rollin,
A.Vasella,
B.Henrissat.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Myrosinase, an S-glycosidase, hydrolyzes plant anionic 1-thio-beta-d-glucosides
(glucosinolates) considered part of the plant defense system. Although
O-glycosidases are ubiquitous, myrosinase is the only known S-glycosidase. Its
active site is very similar to that of retaining O-glycosidases, but one of the
catalytic residues in O-glycosidases, a carboxylate residue functioning as the
general base, is replaced by a glutamine residue. Myrosinase is strongly
activated by ascorbic acid. Several binary and ternary complexes of myrosinase
with different transition state analogues and ascorbic acid have been analyzed
at high resolution by x-ray crystallography along with a
2-deoxy-2-fluoro-glucosyl enzyme intermediate. One of the inhibitors,
d-gluconhydroximo-1,5-lactam, binds simultaneously with a sulfate ion to form a
mimic of the enzyme-substrate complex. Ascorbate binds to a site distinct from
the glucose binding site but overlapping with the aglycon binding site,
suggesting that activation occurs at the second step of catalysis, i.e.
hydrolysis of the glycosyl enzyme. A water molecule is placed perfectly for
activation by ascorbate and for nucleophilic attack on the covalently trapped
2-fluoro-glucosyl-moiety. Activation of the hydrolysis of the glucosyl enzyme
intermediate is further evidenced by the observation that ascorbate enhances the
rate of reactivation of the 2-fluoro-glycosyl enzyme, leading to the conclusion
that ascorbic acid substitutes for the catalytic base in myrosinase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. The binding of ascorbate. The structures have
been obtained on crystals soaked with ascorbic acid and
different inhibitors. Electron density maps as described for
Fig. 2. Water molecules are shown as red spheres. The refined
structures are shown, including ascorbate, water molecules,
sulfate ions, glycerol, inhibitors, and active site residues.
Hydrogen bonds involved in ascorbate recognition are shown as
dotted lines. a, soak with ascorbate, the glycerol molecule
comes from the cryoprotectant. b, ascorbate and
gluco-hydroximolactam. The ascorbate competes with the sulfate
ion that has both partial occupancies of 0.6 for the ascorbate
and 0.4 for the sulfate. c, ascorbate bound to the 2-F-glucosyl
enzyme. The water molecule that is activated by the ascorbate
for an attack on the C-1 carbon of the glucose is indicated by a
pink arrow.
|
 |
Figure 8.
Fig. 8. Schematic reaction mechanism of myrosinase in the
absence (a) and presence (b and c) of ascorbic acid. E, enzyme;
S, substrate; GE, glucosyl enzyme; A, ascorbate; k[1], k[2],
etc., dissociation constants not involving ascorbate; k[3'],
rate constant in presence of ascorbate; k[A1], k[A  1],
etc., dissociation constants of ascorbate. The less important
back reactions are not shown. 1],
etc., dissociation constants of ascorbate. The less important
back reactions are not shown.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2000,
275,
39385-39393)
copyright 2000.
|
|
| |
Figures were
selected
by the author.
|
|

|
| |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|

|
| |
The enzyme is a dimer linked in particular through a Zn2+ ion (ZN1502) located on the 2-fold dimer axis and coordinated by 2 aspartic acid (ASP M70)and 2 histidine residues (HIS M56).
The full heptasaccharide typical for plant glycosylation is visible attached to ASN M292.
Wim Burmeister
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Nong,
J.M.Zhang,
D.Q.Li,
M.Wang,
X.P.Sun,
Y.J.Zhu,
J.Meijer,
and
Q.H.Wang
(2010).
Characterization of a novel β-thioglucosidase CpTGG1 in Carica papaya and its substrate-dependent and ascorbic acid-independent O-β-glucosidase activity.
|
| |
J Integr Plant Biol,
52,
879-890.
|
 |
|
|
|
|
 |
J.R.Ketudat Cairns,
and
A.Esen
(2010).
β-Glucosidases.
|
| |
Cell Mol Life Sci,
67,
3389-3405.
|
 |
|
|
|
|
 |
P.J.Turnbaugh,
B.Henrissat,
and
J.I.Gordon
(2010).
Viewing the human microbiome through three-dimensional glasses: integrating structural and functional studies to better define the properties of myriad carbohydrate-active enzymes.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1261-1264.
|
 |
|
|
|
|
 |
T.M.Gloster,
and
G.J.Davies
(2010).
Glycosidase inhibition: assessing mimicry of the transition state.
|
| |
Org Biomol Chem,
8,
305-320.
|
 |
|
|
|
|
 |
T.V.Vuong,
and
D.B.Wilson
(2010).
Glycoside hydrolases: catalytic base/nucleophile diversity.
|
| |
Biotechnol Bioeng,
107,
195-205.
|
 |
|
|
|
|
 |
W.P.Suza,
C.A.Avila,
K.Carruthers,
S.Kulkarni,
F.L.Goggin,
and
A.Lorence
(2010).
Exploring the impact of wounding and jasmonates on ascorbate metabolism.
|
| |
Plant Physiol Biochem,
48,
337-350.
|
 |
|
|
|
|
 |
N.K.Clay,
A.M.Adio,
C.Denoux,
G.Jander,
and
F.M.Ausubel
(2009).
Glucosinolate metabolites required for an Arabidopsis innate immune response.
|
| |
Science,
323,
95.
|
 |
|
|
|
|
 |
A.D.Hill,
and
P.J.Reilly
(2008).
A Gibbs free energy correlation for automated docking of carbohydrates.
|
| |
J Comput Chem,
29,
1131-1141.
|
 |
|
|
|
|
 |
A.D.Hill,
and
P.J.Reilly
(2008).
Computational analysis of glycoside hydrolase family 1 specificities.
|
| |
Biopolymers,
89,
1021-1031.
|
 |
|
|
|
|
 |
C.Jacob,
and
A.Anwar
(2008).
The chemistry behind redox regulation with a focus on sulphur redox systems.
|
| |
Physiol Plant,
133,
469-480.
|
 |
|
|
|
|
 |
K.Schlaeppi,
N.Bodenhausen,
A.Buchala,
F.Mauch,
and
P.Reymond
(2008).
The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis.
|
| |
Plant J,
55,
774-786.
|
 |
|
|
|
|
 |
B.A.Halkier,
and
J.Gershenzon
(2006).
Biology and biochemistry of glucosinolates.
|
| |
Annu Rev Plant Biol,
57,
303-333.
|
 |
|
|
|
|
 |
C.D.Grubb,
and
S.Abel
(2006).
Glucosinolate metabolism and its control.
|
| |
Trends Plant Sci,
11,
89.
|
 |
|
|
|
|
 |
K.A.Nielsen,
M.Hrmova,
J.N.Nielsen,
K.Forslund,
S.Ebert,
C.E.Olsen,
G.B.Fincher,
and
B.L.Møller
(2006).
Reconstitution of cyanogenesis in barley (Hordeum vulgare L.) and its implications for resistance against the barley powdery mildew fungus.
|
| |
Planta,
223,
1010-1023.
|
 |
|
|
|
|
 |
M.Burow,
J.Markert,
J.Gershenzon,
and
U.Wittstock
(2006).
Comparative biochemical characterization of nitrile-forming proteins from plants and insects that alter myrosinase-catalysed hydrolysis of glucosinolates.
|
| |
FEBS J,
273,
2432-2446.
|
 |
|
|
|
|
 |
P.J.Linley,
M.Landsberger,
T.Kohchi,
J.B.Cooper,
and
M.J.Terry
(2006).
The molecular basis of heme oxygenase deficiency in the pcd1 mutant of pea.
|
| |
FEBS J,
273,
2594-2606.
|
 |
|
|
|
|
 |
A.Bourderioux,
M.Lefoix,
D.Gueyrard,
A.Tatibouét,
S.Cottaz,
S.Arzt,
W.P.Burmeister,
and
P.Rollin
(2005).
The glucosinolate-myrosinase system. New insights into enzyme-substrate interactions by use of simplified inhibitors.
|
| |
Org Biomol Chem,
3,
1872-1879.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Vasella,
G.J.Davies,
and
M.Böhm
(2002).
Glycosidase mechanisms.
|
| |
Curr Opin Chem Biol,
6,
619-629.
|
 |
|
|
|
|
 |
D.L.Zechel,
and
S.G.Withers
(2001).
Dissection of nucleophilic and acid-base catalysis in glycosidases.
|
| |
Curr Opin Chem Biol,
5,
643-649.
|
 |
|
|
|
|
 |
Y.Bourne,
and
B.Henrissat
(2001).
Glycoside hydrolases and glycosyltransferases: families and functional modules.
|
| |
Curr Opin Struct Biol,
11,
593-600.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

