 |
PDBsum entry 1e4b

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Aldolase (class ii)
|
PDB id
|
|

|

|
1e4b
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.17
- L-fuculose-phosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-fuculose 1-phosphate = (S)-lactaldehyde + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
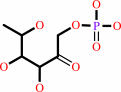 L-fuculose 1-phosphate
L-fuculose 1-phosphate
|
=
|
(S)-lactaldehyde
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
303:531-543
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of l-fuculose-1-phosphate aldolase mutants outlining motions during catalysis.
|
|
A.C.Joerger,
C.Mueller-Dieckmann,
G.E.Schulz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of l-fuculose-1-phosphate aldolase (FucA) with and
without a ligated analogue of dihydroxyacetone phosphate (DHAP) and of a number
of active center mutants have resulted in a model of the catalytic mechanism.
This model has now been confirmed by structural analyses of further mutations at
the zinc coordination sphere and at the phosphate site. In addition, these
mutants have revealed new aspects of the catalysis: the hydroxyl group of
Tyr113' (from a neighboring subunit), which sits just outside the zinc
coordination sphere, steers DHAP towards a productive binding mode at the zinc
ion; Glu73 contacts zinc in between the two ligand positions intended for the
DHAP oxygen atoms and thus avoids blocking of these positions by a tetrahedrally
coordinated hydroxy ion; the FucA polypeptide does not assume its minimum energy
state but oscillates between two states of elevated energy as demonstrated by a
mutant in a minimum energy state. The back and forth motion involves a mobile
loop connecting the phosphate site with intersubunit motions and thus with the
Brownian motion of the solvent. The phosphate group is bound strongly at a given
distance to the zinc ion, which prevents the formation of too tight a DHAP:zinc
complex. This observation explains our failure to find mutants that accept
phosphate-free substitutes for DHAP. The FucA zinc coordination sphere is
compared with that of carbonic anhydrase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Stereo views of the active center of FucA: (a)
the complex FucA:Sulf with a sulfate ion from the
crystallization buffer bound at the phosphate site [Dreyer and
Schulz 1993 and Dreyer and Schulz 1996a] containing the mobile
loop (23-27) (orange); (b) the complex FucA:PGH with the
transition state analogue phosphoglycolohydroxamate [Dreyer and
Schulz 1996b] showing the solidified loop (23-27) and including
a model (orange) of bound Image -lactaldehyde and solidified
Tyr209' [Joerger et al 2000].
|
 |
Figure 7.
Figure 7. The zinc coordination spheres of FucA:Sulf
(yellow), FucA:PGH (red), FucA mutants Glu73->Ser (blue),
Glu73->Gln (light blue/grey), E73Q/Y113F/Y209F (magenta),
Ser71->Gln (green) and of carbonic anhydrase (pink, tetrahedron
drawn out) [Hakansson et al 1992]. The structures were
superimposed on the contacting histidine nitrogen atoms and the
zinc ion. The N  epsilon,
Greek atoms of histidine 92, 94 and 155 and the zinc ion of FucA
are shown in the respective colors. They correspond to His96-N epsilon,
Greek atoms of histidine 92, 94 and 155 and the zinc ion of FucA
are shown in the respective colors. They correspond to His96-N
 epsilon,
Greek , His94-N epsilon,
Greek , His94-N  epsilon,
Greek , and His119-Nd and the zinc ion of carbonic anhydrase,
respectively. epsilon,
Greek , and His119-Nd and the zinc ion of carbonic anhydrase,
respectively.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
303,
531-543)
copyright 2000.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
X.Garrabou,
L.Gómez,
J.Joglar,
S.Gil,
T.Parella,
J.Bujons,
and
P.Clapés
(2010).
Structure-guided minimalist redesign of the L-fuculose-1-phosphate aldolase active site: expedient synthesis of novel polyhydroxylated pyrrolizidines and their inhibitory properties against glycosidases and intestinal disaccharidases.
|
| |
Chemistry,
16,
10691-10706.
|
 |
|
|
|
|
 |
H.Ashida,
Y.Saito,
C.Kojima,
and
A.Yokota
(2008).
Enzymatic characterization of 5-methylthioribulose-1-phosphate dehydratase of the methionine salvage pathway in Bacillus subtilis.
|
| |
Biosci Biotechnol Biochem,
72,
959-967.
|
 |
|
|
|
|
 |
S.Keller,
F.Pojer,
L.Heide,
and
D.M.Lawson
(2006).
Molecular replacement in the 'twilight zone': structure determination of the non-haem iron oxygenase NovR from Streptomyces spheroides through repeated density modification of a poor molecular-replacement solution.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1564-1570.
|
 |
|
|
|
|
 |
L.Espelt,
J.Bujons,
T.Parella,
J.Calveras,
J.Joglar,
A.Delgado,
and
P.Clapés
(2005).
Aldol additions of dihydroxyacetone phosphate to N-Cbz-amino aldehydes catalyzed by L-fuculose-1-phosphate aldolase in emulsion systems: inversion of stereoselectivity as a function of the acceptor aldehyde.
|
| |
Chemistry,
11,
1392-1401.
|
 |
|
|
|
|
 |
M.Kroemer,
and
G.E.Schulz
(2002).
The structure of L-rhamnulose-1-phosphate aldolase (class II) solved by low-resolution SIR phasing and 20-fold NCS averaging.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
824-832.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

