 |
PDBsum entry 1e22

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.6
- lysine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Lys) + L-lysine + ATP = L-lysyl-tRNA(Lys) + AMP + diphosphate
|
 |
 |
 |
 |
 |
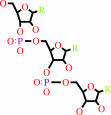 tRNA(Lys)
tRNA(Lys)
|
+
|
L-lysine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
L-lysyl-tRNA(Lys)
Bound ligand (Het Group name = )
matches with 74.19% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:8418-8425
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Active site of lysyl-tRNA synthetase: structural studies of the adenylation reaction.
|
|
G.Desogus,
F.Todone,
P.Brick,
S.Onesti.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aminoacyl-tRNA synthetases play a key role in protein biosynthesis by catalyzing
the specific aminoacylation of tRNA. The energy required for the formation of
the ester bond between the amino acid carboxylate group and the tRNA acceptor
stem is supplied by coupling the reaction to the hydrolysis of ATP. Lysyl-tRNA
synthetase from Escherichia coli belongs to the family of class II synthetases
and carries out a two-step reaction, in which lysine is activated by being
attached to the alpha-phosphate of AMP before being transferred to the cognate
tRNA. Crystals of the thermo-inducible E. coli lysyl-tRNA synthetase LysU which
diffract to 2.1 A resolution have been used to determine crystal structures of
the enzyme in the presence of lysine, the lysyl-adenylate intermediate, and the
nonhydrolyzable ATP analogue AMP-PCP. Additional data have been obtained from
crystals soaked in a solution containing ATP and Mn(2+). The refined crystal
structures give "snapshots" of the active site corresponding to key steps in the
aminoacylation reaction and provide the structural framework for understanding
the mechanism of lysine activation. The active site of LysU is shaped to
position the substrates for the nucleophilic attack of the lysine carboxylate on
the ATP alpha-phosphate. No residues are directly involved in catalysis, but a
number of highly conserved amino acids and three metal ions coordinate the
substrates and stabilize the pentavalent transition state. A loop close to the
catalytic pocket, disordered in the lysine-bound structure, becomes ordered upon
adenine binding.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.V.Palanivelu,
A.Goepfert,
M.Meury,
P.Guye,
C.Dehio,
and
T.Schirmer
(2011).
Fic domain-catalyzed adenylylation: insight provided by the structural analysis of the type IV secretion system effector BepA.
|
| |
Protein Sci,
20,
492-499.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Yanagisawa,
T.Sumida,
R.Ishii,
C.Takemoto,
and
S.Yokoyama
(2010).
A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
|
| |
Nat Struct Mol Biol,
17,
1136-1143.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.W.Navarre,
S.B.Zou,
H.Roy,
J.L.Xie,
A.Savchenko,
A.Singer,
E.Edvokimova,
L.R.Prost,
R.Kumar,
M.Ibba,
and
F.C.Fang
(2010).
PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica.
|
| |
Mol Cell,
39,
209-221.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Fan,
and
J.S.Blanchard
(2009).
Toward the catalytic mechanism of a cysteine ligase (MshC) from Mycobacterium smegmatis: an enzyme involved in the biosynthetic pathway of mycothiol.
|
| |
Biochemistry,
48,
7150-7159.
|
 |
|
|
|
|
 |
H.Sakurama,
T.Takita,
B.Mikami,
T.Itoh,
K.Yasukawa,
and
K.Inouye
(2009).
Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate.
|
| |
J Biochem,
145,
555-563.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Thompson,
C.Lazennec,
P.Plateau,
and
T.Simonson
(2008).
Probing electrostatic interactions and ligand binding in aspartyl-tRNA synthetase through site-directed mutagenesis and computer simulations.
|
| |
Proteins,
71,
1450-1460.
|
 |
|
|
|
|
 |
N.Shen,
M.Zhou,
B.Yang,
Y.Yu,
X.Dong,
and
J.Ding
(2008).
Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states.
|
| |
Nucleic Acids Res,
36,
1288-1299.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Yanagisawa,
R.Ishii,
R.Fukunaga,
T.Kobayashi,
K.Sakamoto,
and
S.Yokoyama
(2008).
Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification.
|
| |
Chem Biol,
15,
1187-1197.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Thompson,
C.Lazennec,
P.Plateau,
and
T.Simonson
(2007).
Ammonium scanning in an enzyme active site. The chiral specificity of aspartyl-tRNA synthetase.
|
| |
J Biol Chem,
282,
30856-30868.
|
 |
|
|
|
|
 |
J.M.Kavran,
S.Gundllapalli,
P.O'Donoghue,
M.Englert,
D.Söll,
and
T.A.Steitz
(2007).
Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
|
| |
Proc Natl Acad Sci U S A,
104,
11268-11273.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Kuratani,
Y.Yoshikawa,
Y.Bessho,
K.Higashijima,
T.Ishii,
R.Shibata,
S.Takahashi,
K.Yutani,
and
S.Yokoyama
(2007).
Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine.
|
| |
Structure,
15,
1642-1653.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Retailleau,
V.Weinreb,
M.Hu,
and
C.W.Carter
(2007).
Crystal structure of tryptophanyl-tRNA synthetase complexed with adenosine-5' tetraphosphate: evidence for distributed use of catalytic binding energy in amino acid activation by class I aminoacyl-tRNA synthetases.
|
| |
J Mol Biol,
369,
108-128.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.F.Chou,
and
C.R.Wagner
(2007).
Lysyl-tRNA synthetase-generated lysyl-adenylate is a substrate for histidine triad nucleotide binding proteins.
|
| |
J Biol Chem,
282,
4719-4727.
|
 |
|
|
|
|
 |
D.Thompson,
P.Plateau,
and
T.Simonson
(2006).
Free-energy simulations and experiments reveal long-range electrostatic interactions and substrate-assisted specificity in an aminoacyl-tRNA synthetase.
|
| |
Chembiochem,
7,
337-344.
|
 |
|
|
|
|
 |
D.Thompson,
and
T.Simonson
(2006).
Molecular dynamics simulations show that bound Mg2+ contributes to amino acid and aminoacyl adenylate binding specificity in aspartyl-tRNA synthetase through long range electrostatic interactions.
|
| |
J Biol Chem,
281,
23792-23803.
|
 |
|
|
|
|
 |
S.J.Hughes,
J.A.Tanner,
A.D.Miller,
and
I.R.Gould
(2006).
Molecular dynamics simulations of LysRS: an asymmetric state.
|
| |
Proteins,
62,
649-662.
|
 |
|
|
|
|
 |
Z.Tokgöz,
R.N.Bohnsack,
and
A.L.Haas
(2006).
Pleiotropic effects of ATP.Mg2+ binding in the catalytic cycle of ubiquitin-activating enzyme.
|
| |
J Biol Chem,
281,
14729-14737.
|
 |
|
|
|
|
 |
M.A.Swairjo,
and
P.R.Schimmel
(2005).
Breaking sieve for steric exclusion of a noncognate amino acid from active site of a tRNA synthetase.
|
| |
Proc Natl Acad Sci U S A,
102,
988-993.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Charron,
H.Roy,
M.Blaise,
R.Giegé,
and
D.Kern
(2003).
Non-discriminating and discriminating aspartyl-tRNA synthetases differ in the anticodon-binding domain.
|
| |
EMBO J,
22,
1632-1643.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.L.Bovee,
M.A.Pierce,
and
C.S.Francklyn
(2003).
Induced fit and kinetic mechanism of adenylation catalyzed by Escherichia coli threonyl-tRNA synthetase.
|
| |
Biochemistry,
42,
15102-15113.
|
 |
|
|
|
|
 |
S.J.Hughes,
J.A.Tanner,
A.D.Hindley,
A.D.Miller,
and
I.R.Gould
(2003).
Functional asymmetry in the lysyl-tRNA synthetase explored by molecular dynamics, free energy calculations and experiment.
|
| |
BMC Struct Biol,
3,
5.
|
 |
|
|
|
|
 |
C.T.Lemke,
and
P.L.Howell
(2002).
Substrate induced conformational changes in argininosuccinate synthetase.
|
| |
J Biol Chem,
277,
13074-13081.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Srinivasan,
C.M.James,
and
J.A.Krzycki
(2002).
Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA.
|
| |
Science,
296,
1459-1462.
|
 |
|
|
|
|
 |
J.A.Tanner,
A.Abowath,
and
A.D.Miller
(2002).
Isothermal titration calorimetry reveals a zinc ion as an atomic switch in the diadenosine polyphosphates.
|
| |
J Biol Chem,
277,
3073-3078.
|
 |
|
|
|
|
 |
J.J.May,
N.Kessler,
M.A.Marahiel,
and
M.T.Stubbs
(2002).
Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
|
| |
Proc Natl Acad Sci U S A,
99,
12120-12125.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Takita,
and
K.Inouye
(2002).
Transition state stabilization by the N-terminal anticodon-binding domain of lysyl-tRNA synthetase.
|
| |
J Biol Chem,
277,
29275-29282.
|
 |
|
|
|
|
 |
R.Fishman,
V.Ankilova,
N.Moor,
and
M.Safro
(2001).
Structure at 2.6 A resolution of phenylalanyl-tRNA synthetase complexed with phenylalanyl-adenylate in the presence of manganese.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1534-1544.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Onesti,
G.Desogus,
A.Brevet,
J.Chen,
P.Plateau,
S.Blanquet,
and
P.Brick
(2000).
Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
|
| |
Biochemistry,
39,
12853-12861.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

