 |
PDBsum entry 1e0c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Sulfurtransferase
|
PDB id
|
|

|

|
1e0c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.8.1.1
- thiosulfate sulfurtransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
thiosulfate + hydrogen cyanide = thiocyanate + sulfite + 2 H+
|
 |
 |
 |
 |
 |
 thiosulfate
thiosulfate
|
+
|
 hydrogen cyanide
hydrogen cyanide
|
=
|
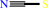 thiocyanate
thiocyanate
|
+
|
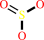 sulfite
sulfite
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
298:691-704
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of a sulfurtransferase from Azotobacter vinelandii highlights the evolutionary relationship between the rhodanese and phosphatase enzyme families.
|
|
D.Bordo,
D.Deriu,
R.Colnaghi,
A.Carpen,
S.Pagani,
M.Bolognesi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Rhodanese is an ubiquitous enzyme that in vitro catalyses the transfer of a
sulfur atom from suitable donors to nucleophilic acceptors by way of a double
displacement mechanism. During the catalytic process the enzyme cycles between a
sulfur-free and a persulfide-containing form, via formation of a persulfide
linkage to a catalytic Cys residue. In the nitrogen-fixing bacteria Azotobacter
vinelandii the rhdA gene has been identified and the encoded protein
functionally characterized as a rhodanese. The crystal structure of the A.
vinelandii rhodanese has been determined and refined at 1.8 A resolution in the
sulfur-free and persulfide-containing forms. Conservation of the overall
three-dimensional fold of bovine rhodanese is observed, with substantial
modifications of the protein structure in the proximity of the catalytic residue
Cys230. Remarkably, the native enzyme is found as the Cys230-persulfide form; in
the sulfur-free state the catalytic Cys residue adopts two alternate
conformations, reflected by perturbation of the neighboring active-site
residues, which is associated with a partly reversible loss of
thiosulfate:cyanide sulfurtransferase activity. The catalytic mechanism of A.
vinelandii rhodanese relies primarily on the main-chain conformation of the 230
to 235 active-site loop and on a surrounding strong positive electrostatic
field. Substrate recognition is based on residues which are entirely different
in the prokaryotic and eukaryotic enzymes. The active-site loop of A. vinelandii
rhodanese displays striking structural similarity to the active-site loop of the
similarly folded catalytic domain of dual specific phosphatase Cdc25, suggesting
a common evolutionary origin of the two enzyme families.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Overall structure of A. vinelandii rhodanese. (a)
Stereoview of the C^a trace, with the molecular pseudo 2-fold
axis approximately normal to the plane of the image. (b) Ribbon
representation of RhdA. The N- and C-terminal domains (brown and
green, respectively), the linker peptide (blue). The secondary
structure elements of each domain are labeled with letters
following the scheme proposed for bovine rhodanese [Ploegman et
al 1978]. A single quote indicates elements of the C-terminal
domains. The active-site loop is shown in red; the catalytic
residue, Cys230, is represented in ball and stick. The drawings
were prepared with the programs MOLSCRIPT [Kraulis 1991] and
Raster3D [Merrit and Murphy 1994].
|
 |
Figure 3.
Figure 3. Stereo representation of the active-site
environment of the sulfur-free rhodanese. The alternate
conformations of Cys230, Arg235, and Trp195 side-chains are
shown in grey and green, respectively; hydrogen bonds as red
dotted lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
298,
691-704)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.Cartini,
W.Remelli,
P.C.Dos Santos,
J.Papenbrock,
S.Pagani,
and
F.Forlani
(2011).
Mobilization of sulfane sulfur from cysteine desulfurases to the Azotobacter vinelandii sulfurtransferase RhdA.
|
| |
Amino Acids,
41,
141-150.
|
 |
|
|
|
|
 |
J.Papenbrock,
S.Guretzki,
and
M.Henne
(2011).
Latest news about the sulfurtransferase protein family of higher plants.
|
| |
Amino Acids,
41,
43-57.
|
 |
|
|
|
|
 |
R.Shi,
A.Proteau,
M.Villarroya,
I.Moukadiri,
L.Zhang,
J.F.Trempe,
A.Matte,
M.E.Armengod,
and
M.Cygler
(2010).
Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
|
| |
PLoS Biol,
8,
e1000354.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Remelli,
A.Cereda,
J.Papenbrock,
F.Forlani,
and
S.Pagani
(2010).
The rhodanese RhdA helps Azotobacter vinelandii in maintaining cellular redox balance.
|
| |
Biol Chem,
391,
777-784.
|
 |
|
|
|
|
 |
H.K.Yeo,
and
J.Y.Lee
(2009).
Crystal structure of Saccharomyces cerevisiae Ygr203w, a homolog of single-domain rhodanese and Cdc25 phosphatase catalytic domain.
|
| |
Proteins,
76,
520-524.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.R.Wallen,
T.C.Mallett,
W.Boles,
D.Parsonage,
C.M.Furdui,
P.A.Karplus,
and
A.Claiborne
(2009).
Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
|
| |
Biochemistry,
48,
9650-9667.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Hänzelmann,
J.U.Dahl,
J.Kuper,
A.Urban,
U.Müller-Theissen,
S.Leimkühler,
and
H.Schindelin
(2009).
Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
|
| |
Protein Sci,
18,
2480-2491.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Cheng,
J.L.Donahue,
S.E.Battle,
W.K.Ray,
and
T.J.Larson
(2008).
Biochemical and Genetic Characterization of PspE and GlpE, Two Single-domain Sulfurtransferases of Escherichia coli.
|
| |
Open Microbiol J,
2,
18-28.
|
 |
|
|
|
|
 |
R.Sabelli,
E.Iorio,
A.De Martino,
F.Podo,
A.Ricci,
G.Viticchiè,
G.Rotilio,
M.Paci,
and
S.Melino
(2008).
Rhodanese-thioredoxin system and allyl sulfur compounds.
|
| |
FEBS J,
275,
3884-3899.
|
 |
|
|
|
|
 |
A.Bartels,
F.Forlani,
S.Pagani,
and
J.Papenbrock
(2007).
Conformational studies on Arabidopsis sulfurtransferase AtStr1 with spectroscopic methods.
|
| |
Biol Chem,
388,
53-59.
|
 |
|
|
|
|
 |
L.Cavalca,
N.Guerrieri,
M.Colombo,
S.Pagani,
and
V.Andreoni
(2007).
Enzymatic and genetic profiles in environmental strains grown on polycyclic aromatic hydrocarbons.
|
| |
Antonie Van Leeuwenhoek,
91,
315-325.
|
 |
|
|
|
|
 |
M.C.Giuliani,
P.Tron,
G.Leroy,
C.Aubert,
P.Tauc,
and
M.T.Giudici-Orticoni
(2007).
A new sulfurtransferase from the hyperthermophilic bacterium Aquifex aeolicus. Being single is not so simple when temperature gets high.
|
| |
FEBS J,
274,
4572-4587.
|
 |
|
|
|
|
 |
V.Sauvé,
S.Bruno,
B.C.Berks,
and
A.M.Hemmings
(2007).
The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm.
|
| |
J Biol Chem,
282,
23194-23204.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Tao,
and
L.Tong
(2007).
Crystal structure of the MAP kinase binding domain and the catalytic domain of human MKP5.
|
| |
Protein Sci,
16,
880-886.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Bisacchi,
Y.Zhou,
B.P.Rosen,
R.Mukhopadhyay,
and
D.Bordo
(2006).
Crystallization and preliminary crystallographic characterization of LmACR2, an arsenate/antimonate reductase from Leishmania major.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
976-979.
|
 |
|
|
|
|
 |
M.Hattori,
E.Mizohata,
A.Tatsuguchi,
R.Shibata,
S.Kishishita,
K.Murayama,
T.Terada,
S.Kuramitsu,
M.Shirouzu,
and
S.Yokoyama
(2006).
Crystal structure of the single-domain rhodanese homologue TTHA0613 from Thermus thermophilus HB8.
|
| |
Proteins,
64,
284-287.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Urich,
C.M.Gomes,
A.Kletzin,
and
C.Frazão
(2006).
X-ray Structure of a self-compartmentalizing sulfur cycle metalloenzyme.
|
| |
Science,
311,
996.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Prokop,
F.Oplustil,
J.DeFrank,
and
J.Damborský
(2006).
Enzymes fight chemical weapons.
|
| |
Biotechnol J,
1,
1370-1380.
|
 |
|
|
|
|
 |
D.Pantoja-Uceda,
B.López-Méndez,
S.Koshiba,
M.Inoue,
T.Kigawa,
T.Terada,
M.Shirouzu,
A.Tanaka,
M.Seki,
K.Shinozaki,
S.Yokoyama,
and
P.Güntert
(2005).
Solution structure of the rhodanese homology domain At4g01050(175-295) from Arabidopsis thaliana.
|
| |
Protein Sci,
14,
224-230.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.S.Carroll,
H.Gao,
H.Chen,
C.D.Stout,
J.A.Leary,
and
C.R.Bertozzi
(2005).
A conserved mechanism for sulfonucleotide reduction.
|
| |
PLoS Biol,
3,
e250.
|
 |
|
|
|
|
 |
M.Acosta,
S.Beard,
J.Ponce,
M.Vera,
J.C.Mobarec,
and
C.A.Jerez
(2005).
Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: structural and functional characterization of the proteins.
|
| |
OMICS,
9,
13-29.
|
 |
|
|
|
|
 |
A.Cereda,
F.Forlani,
S.Iametti,
R.Bernhardt,
P.Ferranti,
G.Picariello,
S.Pagani,
and
F.Bonomi
(2003).
Molecular recognition between Azotobacter vinelandii rhodanese and a sulfur acceptor protein.
|
| |
Biol Chem,
384,
1473-1481.
|
 |
|
|
|
|
 |
R.A.Williams,
S.M.Kelly,
J.C.Mottram,
and
G.H.Coombs
(2003).
3-Mercaptopyruvate sulfurtransferase of Leishmania contains an unusual C-terminal extension and is involved in thioredoxin and antioxidant metabolism.
|
| |
J Biol Chem,
278,
1480-1486.
|
 |
|
|
|
|
 |
S.Melino,
D.O.Cicero,
M.Orsale,
F.Forlani,
S.Pagani,
and
M.Paci
(2003).
Azotobacter vinelandii rhodanese: selenium loading and ion interaction studies.
|
| |
Eur J Biochem,
270,
4208-4215.
|
 |
|
|
|
|
 |
A.E.Todd,
C.A.Orengo,
and
J.M.Thornton
(2002).
Sequence and structural differences between enzyme and nonenzyme homologs.
|
| |
Structure,
10,
1435-1451.
|
 |
|
|
|
|
 |
D.Bordo,
and
P.Bork
(2002).
The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations.
|
| |
EMBO Rep,
3,
741-746.
|
 |
|
|
|
|
 |
M.Burow,
D.Kessler,
and
J.Papenbrock
(2002).
Enzymatic activity of the Arabidopsis sulfurtransferase resides in the C-terminal domain but is boosted by the N-terminal domain and the linker peptide in the full-length enzyme.
|
| |
Biol Chem,
383,
1363-1372.
|
 |
|
|
|
|
 |
V.A.Bamford,
S.Bruno,
T.Rasmussen,
C.Appia-Ayme,
M.R.Cheesman,
B.C.Berks,
and
A.M.Hemmings
(2002).
Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme.
|
| |
EMBO J,
21,
5599-5610.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Spallarossa,
J.L.Donahue,
T.J.Larson,
M.Bolognesi,
and
D.Bordo
(2001).
Escherichia coli GlpE is a prototype sulfurtransferase for the single-domain rhodanese homology superfamily.
|
| |
Structure,
9,
1117-1125.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Bordo,
F.Forlani,
A.Spallarossa,
R.Colnaghi,
A.Carpen,
M.Bolognesi,
and
S.Pagani
(2001).
A persulfurated cysteine promotes active site reactivity in Azotobacter vinelandii Rhodanese.
|
| |
Biol Chem,
382,
1245-1252.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Iametti,
A.K.Bera,
G.Vecchio,
A.Grinberg,
R.Bernhardt,
and
F.Bonomi
(2001).
GroEL-assisted refolding of adrenodoxin during chemical cluster insertion.
|
| |
Eur J Biochem,
268,
2421-2429.
|
 |
|
|
|
|
 |
D.Bordo,
T.J.Larson,
J.L.Donahue,
A.Spallarossa,
and
M.Bolognesi
(2000).
Crystals of GlpE, a 12 kDa sulfurtransferase from escherichia coli, display 1.06 A resolution diffraction: a preliminary report.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
1691-1693.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

