 |
PDBsum entry 1dob

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1dob
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.13.2
- 4-hydroxybenzoate 3-monooxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxybenzoate + NADPH + O2 + H+ = 3,4-dihydroxybenzoate + NADP+ + H2O
|
 |
 |
 |
 |
 |
4-hydroxybenzoate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADPH
NADPH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
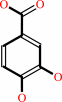 3,4-dihydroxybenzoate
3,4-dihydroxybenzoate
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
266:110-114
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
The mobile flavin of 4-OH benzoate hydroxylase.
|
|
D.L.Gatti,
B.A.Palfey,
M.S.Lah,
B.Entsch,
V.Massey,
D.P.Ballou,
M.L.Ludwig.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Para-hydroxybenzoate hydroxylase inserts oxygen into substrates by means of the
labile intermediate, flavin C(4a)-hydroperoxide. This reaction requires
transient isolation of the flavin and substrate from the bulk solvent. Previous
crystal structures have revealed the position of the substrate
para-hydroxybenzoate during oxygenation but not how it enters the active site.
In this study, enzyme structures with the flavin ring displaced relative to the
protein were determined, and it was established that these or similar flavin
conformations also occur in solution. Movement of the flavin appears to be
essential for the translocation of substrates and products into the
solvent-shielded active site during catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Volkers,
G.J.Palm,
M.S.Weiss,
G.D.Wright,
and
W.Hinrichs
(2011).
Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase.
|
| |
FEBS Lett,
585,
1061-1066.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Sah,
and
P.S.Phale
(2011).
1-Naphthol 2-hydroxylase from Pseudomonas sp. strain C6: purification, characterization and chemical modification studies.
|
| |
Biodegradation,
22,
517-526.
|
 |
|
|
|
|
 |
Y.W.Tan,
and
H.Yang
(2011).
Seeing the forest for the trees: fluorescence studies of single enzymes in the context of ensemble experiments.
|
| |
Phys Chem Chem Phys,
13,
1709-1721.
|
 |
|
|
|
|
 |
U.E.Ukaegbu,
A.Kantz,
M.Beaton,
G.T.Gassner,
and
A.C.Rosenzweig
(2010).
Structure and ligand binding properties of the epoxidase component of styrene monooxygenase .
|
| |
Biochemistry,
49,
1678-1688.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.L.Mobley,
and
K.A.Dill
(2009).
Binding of small-molecule ligands to proteins: "what you see" is not always "what you get".
|
| |
Structure,
17,
489-498.
|
 |
|
|
|
|
 |
R.C.Bruckner,
and
M.S.Jorns
(2009).
Spectral and kinetic characterization of intermediates in the aromatization reaction catalyzed by NikD, an unusual amino acid oxidase.
|
| |
Biochemistry,
48,
4455-4465.
|
 |
|
|
|
|
 |
S.D.Pope,
L.L.Chen,
and
V.Stewart
(2009).
Purine utilization by Klebsiella oxytoca M5al: genes for ring-oxidizing and -opening enzymes.
|
| |
J Bacteriol,
191,
1006-1017.
|
 |
|
|
|
|
 |
T.Senda,
M.Senda,
S.Kimura,
and
T.Ishida
(2009).
Redox control of protein conformation in flavoproteins.
|
| |
Antioxid Redox Signal,
11,
1741-1766.
|
 |
|
|
|
|
 |
E.V.Kudryashova,
A.J.Visser,
and
W.J.van Berkel
(2008).
Monomer formation and function of p-hydroxybenzoate hydroxylase in reverse micelles and in dimethylsulfoxide/water mixtures.
|
| |
Chembiochem,
9,
413-419.
|
 |
|
|
|
|
 |
Y.Huang,
K.X.Zhao,
X.H.Shen,
C.Y.Jiang,
and
S.J.Liu
(2008).
Genetic and biochemical characterization of a 4-hydroxybenzoate hydroxylase from Corynebacterium glutamicum.
|
| |
Appl Microbiol Biotechnol,
78,
75-83.
|
 |
|
|
|
|
 |
A.Alfieri,
F.Fersini,
N.Ruangchan,
M.Prongjit,
P.Chaiyen,
and
A.Mattevi
(2007).
Structure of the monooxygenase component of a two-component flavoprotein monooxygenase.
|
| |
Proc Natl Acad Sci U S A,
104,
1177-1182.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.P.Ballou
(2007).
Crystallography gets the jump on the enzymologists.
|
| |
Proc Natl Acad Sci U S A,
104,
15587-15588.
|
 |
|
|
|
|
 |
K.M.Meneely,
and
A.L.Lamb
(2007).
Biochemical characterization of a flavin adenine dinucleotide-dependent monooxygenase, ornithine hydroxylase from Pseudomonas aeruginosa, suggests a novel reaction mechanism.
|
| |
Biochemistry,
46,
11930-11937.
|
 |
|
|
|
|
 |
K.S.Ryan,
A.R.Howard-Jones,
M.J.Hamill,
S.J.Elliott,
C.T.Walsh,
and
C.L.Drennan
(2007).
Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC.
|
| |
Proc Natl Acad Sci U S A,
104,
15311-15316.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.M.Moustafa,
S.Foster,
A.Y.Lyubimov,
and
A.Vrielink
(2006).
Crystal structure of LAAO from Calloselasma rhodostoma with an L-phenylalanine substrate: insights into structure and mechanism.
|
| |
J Mol Biol,
364,
991.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.A.Feenstra,
K.Hofstetter,
R.Bosch,
A.Schmid,
J.N.Commandeur,
and
N.P.Vermeulen
(2006).
Enantioselective substrate binding in a monooxygenase protein model by molecular dynamics and docking.
|
| |
Biophys J,
91,
3206-3216.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Siebold,
N.Berrow,
T.S.Walter,
K.Harlos,
R.J.Owens,
D.I.Stuart,
J.R.Terman,
A.L.Kolodkin,
R.J.Pasterkamp,
and
E.Y.Jones
(2005).
High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule.
|
| |
Proc Natl Acad Sci U S A,
102,
16836-16841.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.M.Young,
D.Parke,
and
L.N.Ornston
(2005).
Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation.
|
| |
Annu Rev Microbiol,
59,
519-551.
|
 |
|
|
|
|
 |
M.Nadella,
M.A.Bianchet,
S.B.Gabelli,
J.Barrila,
and
L.M.Amzel
(2005).
Structure and activity of the axon guidance protein MICAL.
|
| |
Proc Natl Acad Sci U S A,
102,
16830-16835.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Oonanant,
J.Sucharitakul,
J.Yuvaniyama,
and
P.Chaiyen
(2005).
Crystallization and preliminary X-ray crystallographic analysis of 2-methyl-3-hydroxypyridine-5-carboxylic acid (MHPC) oxygenase from Pseudomonas sp. MA-1.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
312-314.
|
 |
|
|
|
|
 |
X.Gao,
C.L.Tan,
C.C.Yeo,
and
C.L.Poh
(2005).
Molecular and biochemical characterization of the xlnD-encoded 3-hydroxybenzoate 6-hydroxylase involved in the degradation of 2,5-xylenol via the gentisate pathway in Pseudomonas alcaligenes NCIMB 9867.
|
| |
J Bacteriol,
187,
7696-7702.
|
 |
|
|
|
|
 |
M.H.Hefti,
J.Vervoort,
and
W.J.van Berkel
(2003).
Deflavination and reconstitution of flavoproteins.
|
| |
Eur J Biochem,
270,
4227-4242.
|
 |
|
|
|
|
 |
B.A.Palfey,
R.Basu,
K.K.Frederick,
B.Entsch,
and
D.P.Ballou
(2002).
Role of protein flexibility in the catalytic cycle of p-hydroxybenzoate hydroxylase elucidated by the Pro293Ser mutant.
|
| |
Biochemistry,
41,
8438-8446.
|
 |
|
|
|
|
 |
C.Sánchez,
I.A.Butovich,
A.F.Braña,
J.Rohr,
C.Méndez,
and
J.A.Salas
(2002).
The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives.
|
| |
Chem Biol,
9,
519-531.
|
 |
|
|
|
|
 |
J.Wang,
M.Ortiz-Maldonado,
B.Entsch,
V.Massey,
D.Ballou,
and
D.L.Gatti
(2002).
Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
|
| |
Proc Natl Acad Sci U S A,
99,
608-613.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.T.Gallagher,
M.Mayhew,
M.J.Holden,
A.Howard,
K.J.Kim,
and
V.L.Vilker
(2001).
The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
|
| |
Proteins,
44,
304-311.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.D.Altose,
Y.Zheng,
J.Dong,
B.A.Palfey,
and
P.R.Carey
(2001).
Comparing protein-ligand interactions in solution and single crystals by Raman spectroscopy.
|
| |
Proc Natl Acad Sci U S A,
98,
3006-3011.
|
 |
|
|
|
|
 |
M.Ortiz-Maldonado,
D.P.Ballou,
and
V.Massey
(2001).
A rate-limiting conformational change of the flavin in p-hydroxybenzoate hydroxylase is necessary for ligand exchange and catalysis: studies with 8-mercapto- and 8-hydroxy-flavins.
|
| |
Biochemistry,
40,
1091-1101.
|
 |
|
|
|
|
 |
R.J.Stanley
(2001).
Advances in flavin and flavoprotein optical spectroscopy.
|
| |
Antioxid Redox Signal,
3,
847-866.
|
 |
|
|
|
|
 |
M.H.Eppink,
E.Cammaart,
D.Van Wassenaar,
W.J.Middelhoven,
and
W.J.van Berkel
(2000).
Purification and properties of hydroquinone hydroxylase, a FAD-dependent monooxygenase involved in the catabolism of 4-hydroxybenzoate in Candida parapsilosis CBS604.
|
| |
Eur J Biochem,
267,
6832-6840.
|
 |
|
|
|
|
 |
D.L.Roberts,
D.Salazar,
J.P.Fulmer,
F.E.Frerman,
and
J.J.Kim
(1999).
Crystal structure of Paracoccus denitrificans electron transfer flavoprotein: structural and electrostatic analysis of a conserved flavin binding domain.
|
| |
Biochemistry,
38,
1977-1989.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.R.Moran,
B.Entsch,
B.A.Palfey,
and
D.P.Ballou
(1999).
Mechanistic insights into p-hydroxybenzoate hydroxylase from studies of the mutant Ser212Ala.
|
| |
Biochemistry,
38,
6292-6299.
|
 |
|
|
|
|
 |
M.Ortiz-Maldonado,
D.Gatti,
D.P.Ballou,
and
V.Massey
(1999).
Structure-function correlations of the reaction of reduced nicotinamide analogues with p-hydroxybenzoate hydroxylase substituted with a series of 8-substituted flavins.
|
| |
Biochemistry,
38,
16636-16647.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ortiz-Maldonado,
D.P.Ballou,
and
V.Massey
(1999).
Use of free energy relationships to probe the individual steps of hydroxylation of p-hydroxybenzoate hydroxylase: studies with a series of 8-substituted flavins.
|
| |
Biochemistry,
38,
8124-8137.
|
 |
|
|
|
|
 |
Y.Zheng,
J.Dong,
B.A.Palfey,
and
P.R.Carey
(1999).
Using Raman spectroscopy to monitor the solvent-exposed and "buried" forms of flavin in p-hydroxybenzoate hydroxylase.
|
| |
Biochemistry,
38,
16727-16732.
|
 |
|
|
|
|
 |
A.Mattevi
(1998).
The PHBH fold: not only flavoenzymes.
|
| |
Biophys Chem,
70,
217-222.
|
 |
|
|
|
|
 |
C.Enroth,
H.Neujahr,
G.Schneider,
and
Y.Lindqvist
(1998).
The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis.
|
| |
Structure,
6,
605-617.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.S.Raman,
H.Li,
P.Martásek,
V.Král,
B.S.Masters,
and
T.L.Poulos
(1998).
Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
|
| |
Cell,
95,
939-950.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.C.Mallett,
and
A.Claiborne
(1998).
Oxygen reactivity of an NADH oxidase C42S mutant: evidence for a C(4a)-peroxyflavin intermediate and a rate-limiting conformational change.
|
| |
Biochemistry,
37,
8790-8802.
|
 |
|
|
|
|
 |
F.J.van der Bolt,
R.H.van den Heuvel,
J.Vervoort,
and
W.J.van Berkel
(1997).
19F NMR study on the regiospecificity of hydroxylation of tetrafluoro-4-hydroxybenzoate by wild-type and Y385F p-hydroxybenzoate hydroxylase: evidence for a consecutive oxygenolytic dehalogenation mechanism.
|
| |
Biochemistry,
36,
14192-14201.
|
 |
|
|
|
|
 |
G.R.Moran,
B.Entsch,
B.A.Palfey,
and
D.P.Ballou
(1997).
Electrostatic effects on substrate activation in para-hydroxybenzoate hydroxylase: studies of the mutant lysine 297 methionine.
|
| |
Biochemistry,
36,
7548-7556.
|
 |
|
|
|
|
 |
M.Tegoni,
M.Gervais,
and
A.Desbois
(1997).
Resonance Raman study on the oxidized and anionic semiquinone forms of flavocytochrome b2 and L-lactate monooxygenase. Influence of the structure and environment of the isoalloxazine ring on the flavin function.
|
| |
Biochemistry,
36,
8932-8946.
|
 |
|
|
|
|
 |
B.Seibold,
M.Matthes,
M.H.Eppink,
F.Lingens,
W.J.Van Berkel,
and
R.Müller
(1996).
4-Hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3. Purification, characterization, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity.
|
| |
Eur J Biochem,
239,
469-478.
|
 |
|
|
|
|
 |
D.L.Gatti,
B.Entsch,
D.P.Ballou,
and
M.L.Ludwig
(1996).
pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
|
| |
Biochemistry,
35,
567-578.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.J.van der Bolt,
J.Vervoort,
and
W.J.van Berkel
(1996).
Flavin motion in p-hydroxybenzoate hydroxylase. Substrate and effector specificity of the Tyr22-->Ala mutant.
|
| |
Eur J Biochem,
237,
592-600.
|
 |
|
|
|
|
 |
G.R.Moran,
B.Entsch,
B.A.Palfey,
and
D.P.Ballou
(1996).
Evidence for flavin movement in the function of p-hydroxybenzoate hydroxylase from studies of the mutant Arg220Lys.
|
| |
Biochemistry,
35,
9278-9285.
|
 |
|
|
|
|
 |
M.H.Eppink,
H.A.Schreuder,
and
W.J.Van Berkel
(1995).
Structure and function of mutant Arg44Lys of 4-hydroxybenzoate hydroxylase implications for NADPH binding.
|
| |
Eur J Biochem,
231,
157-165.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.J.van Berkel,
M.H.Eppink,
and
H.A.Schreuder
(1994).
Crystal structure of p-hydroxybenzoate hydroxylase reconstituted with the modified FAD present in alcohol oxidase from methylotrophic yeasts: evidence for an arabinoflavin.
|
| |
Protein Sci,
3,
2245-2253.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

