 |
PDBsum entry 1dib

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase,hydrolase
|
PDB id
|
|

|

|
1dib
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase,hydrolase
|
 |
|
Title:
|
 |
Human methylenetetrahydrofolate dehydrogenase / cyclohydrolase complexed with NADP and inhibitor ly345899
|
|
Structure:
|
 |
Methylenetetrahydrofolate dehydrogenase/cyclohydrolase. Chain: a, b. Synonym: dc301. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.208
|
R-free:
|
0.240
|
|
|
Authors:
|
 |
A.Schmidt,H.Wu,R.E.Mackenzie,V.J.Chen,J.R.Bewly,J.E.Ray,J.E.Toth, M.Cygler
|
Key ref:

|
 |
A.Schmidt
et al.
(2000).
Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry,
39,
6325-6335.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
29-Nov-99
|
Release date:
|
05-Jul-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P11586
(C1TC_HUMAN) -
C-1-tetrahydrofolate synthase, cytoplasmic from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
935 a.a.
285 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.5.1.5
- methylenetetrahydrofolate dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Folate Coenzymes
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6R)-5,10-methylene-5,6,7,8-tetrahydrofolate + NADP+ = (6R)-5,10- methenyltetrahydrofolate + NADPH
|
 |
 |
 |
 |
 |
(6R)-5,10-methylene-5,6,7,8-tetrahydrofolate
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
(6R)-5,10- methenyltetrahydrofolate
|
+
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.4.9
- methenyltetrahydrofolate cyclohydrolase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6R)-5,10-methenyltetrahydrofolate + H2O = (6R)-10-formyltetrahydrofolate + H+
|
 |
 |
 |
 |
 |
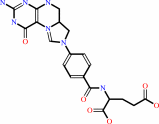 5,10-methenyltetrahydrofolate
5,10-methenyltetrahydrofolate
|
+
|
H2O
|
=
|
(6S)-10-formyltetrahydrofolate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.6.3.4.3
- formate--tetrahydrofolate ligase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6S)-5,6,7,8-tetrahydrofolate + formate + ATP = (6R)-10- formyltetrahydrofolate + ADP + phosphate
|
 |
 |
 |
 |
 |
(6S)-5,6,7,8-tetrahydrofolate
|
+
|
 formate
formate
|
+
|
 ATP
ATP
|
=
|
(6R)-10- formyltetrahydrofolate
|
+
|
ADP
Bound ligand (Het Group name = )
matches with 56.25% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:6325-6335
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
|
|
A.Schmidt,
H.Wu,
R.E.MacKenzie,
V.J.Chen,
J.R.Bewly,
J.E.Ray,
J.E.Toth,
M.Cygler.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Enzymes involved in tetrahydrofolate metabolism are of particular pharmaceutical
interest, as their function is crucial for amino acid and DNA biosynthesis. The
crystal structure of the human cytosolic methylenetetrahydrofolate
dehydrogenase/cyclohydrolase (DC301) domain of a trifunctional enzyme has been
determined previously with a bound NADP cofactor. While the substrate binding
site was identified to be localized in a deep and rather hydrophobic cleft at
the interface between two protein domains, the unambiguous assignment of
catalytic residues was not possible. We succeeded in determining the crystal
structures of three ternary DC301/NADP/inhibitor complexes. Investigation of
these structures followed by site-directed mutagenesis studies allowed
identification of the amino acids involved in catalysis by both enzyme
activities. The inhibitors bind close to Lys56 and Tyr52, residues of a strictly
conserved motif for active sites in dehydrogenases. While Lys56 is in a good
position for chemical interaction with the substrate analogue, Tyr52 was found
stacking against the inhibitors' aromatic rings and hence seems to be more
important for proper positioning of the ligand than for catalysis. Also, Ser49
and/or Cys147 were found to possibly act as an activator for water in the
cyclohydrolase step. These and the other residues (Gln100 and Asp125), with
which contacts are made, are strictly conserved in THF dehydrogenases. On the
basis of structural and mutagenesis data, we propose a reaction mechanism for
both activities, the dehydrogenase and the cyclohydrolase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.N.Lee,
D.Takawira,
A.P.Nikolova,
D.P.Ballou,
V.C.Furtado,
N.L.Phung,
B.R.Still,
M.K.Thorstad,
J.J.Tanner,
and
E.E.Trimmer
(2009).
Functional role for the conformationally mobile phenylalanine 223 in the reaction of methylenetetrahydrofolate reductase from Escherichia coli.
|
| |
Biochemistry,
48,
7673-7685.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Prasannan,
and
D.R.Appling
(2009).
Human mitochondrial C1-tetrahydrofolate synthase: submitochondrial localization of the full-length enzyme and characterization of a short isoform.
|
| |
Arch Biochem Biophys,
481,
86-93.
|
 |
|
|
|
|
 |
S.M.Murta,
T.J.Vickers,
D.A.Scott,
and
S.M.Beverley
(2009).
Methylene tetrahydrofolate dehydrogenase/cyclohydrolase and the synthesis of 10-CHO-THF are essential in Leishmania major.
|
| |
Mol Microbiol,
71,
1386-1401.
|
 |
|
|
|
|
 |
T.J.Vickers,
S.M.Murta,
M.A.Mandell,
and
S.M.Beverley
(2009).
The enzymes of the 10-formyl-tetrahydrofolate synthetic pathway are found exclusively in the cytosol of the trypanosomatid parasite Leishmania major.
|
| |
Mol Biochem Parasitol,
166,
142-152.
|
 |
|
|
|
|
 |
S.W.Ragsdale,
and
E.Pierce
(2008).
Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation.
|
| |
Biochim Biophys Acta,
1784,
1873-1898.
|
 |
|
|
|
|
 |
K.E.Christensen,
H.Patel,
U.Kuzmanov,
N.R.Mejia,
and
R.E.MacKenzie
(2005).
Disruption of the mthfd1 gene reveals a monofunctional 10-formyltetrahydrofolate synthetase in mammalian mitochondria.
|
| |
J Biol Chem,
280,
7597-7602.
|
 |
|
|
|
|
 |
K.E.Christensen,
I.A.Mirza,
A.M.Berghuis,
and
R.E.Mackenzie
(2005).
Magnesium and phosphate ions enable NAD binding to methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase.
|
| |
J Biol Chem,
280,
34316-34323.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Prasannan,
S.Pike,
K.Peng,
B.Shane,
and
D.R.Appling
(2003).
Human mitochondrial C1-tetrahydrofolate synthase: gene structure, tissue distribution of the mRNA, and immunolocalization in Chinese hamster ovary calls.
|
| |
J Biol Chem,
278,
43178-43187.
|
 |
|
|
|
|
 |
S.Sundararajan,
and
R.E.MacKenzie
(2002).
Residues involved in the mechanism of the bifunctional methylenetetrahydrofolate dehydrogenase-cyclohydrolase: the roles of glutamine 100 and aspartate 125.
|
| |
J Biol Chem,
277,
18703-18709.
|
 |
|
|
|
|
 |
U.Ermler,
C.H.Hagemeier,
A.Roth,
U.Demmer,
W.Grabarse,
E.Warkentin,
and
J.A.Vorholt
(2002).
Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
|
| |
Structure,
10,
1127-1137.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

