 |
PDBsum entry 1d9c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Immune system
|
PDB id
|
|

|

|
1d9c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
56:14-24
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
The 2.0 A structure of bovine interferon-gamma; assessment of the structural differences between species.
|
|
M.Randal,
A.A.Kossiakoff.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of bovine interferon-gamma (IFN-gamma) was determined by multiple
isomorphous replacement at 2.0 A resolution. Bovine IFN-gamma crystallizes in
two related crystal forms. Crystal form 1 diffracts to 2.9 A resolution and is
reproducible and stable to derivatization. Crystal form 2 diffracts to 2.0 A
resolution, but shows significant non-isomorphism from crystal to crystal. The
previously determined structures of several different species of INF-gamma were
either at too low a resolution [human, 1hig; Ealick et al. (1991), Science, 252,
or were too inaccurate [bovine, 1rfb; Samudzi & Rubin (1993), Acta
Cryst. D49(6), 505-512; rabbit, 2rig; Samudzi et al. (1991), J. Biol. Chem.
for the structure to be solved by molecular replacement.
The structure was solved in crystal form 1 using two derivatives produced by
chemically modifying two free cysteine residues that were introduced by
site-directed mutagenesis (Ser30Cys, Asn59Cys). After model building and
refinement, the final R value was 21.8% (R(free) = 30.9%) for all data in the
resolution range 8.0-2.9 A. The crystal form 1 structure was then used as a
molecular-replacement model for crystal form 2 data collected from a
flash-cooled crystal. Subsequent model building and refinement, using all data
in the resolution range 15.0-2.0 A, gave an R value of 19.7% and an R(free) of
27.5%. Pairwise comparison of C(alpha) positions of bovine IFN-gamma (BOV) and
the previously determined 1rfb and 2rig structures indicated some significant
differences in the models (r.m.s.d. values for BOV to 1rfb, 4.3 A; BOV to 2rig,
4.0 A). An assessment of the quality of the structures was made using the 3D-1D
algorithm [Eisenberg et al. (1992), Faraday Discuss. 93, 25-34]. The resulting
statistical scoring indicated that BOV was consistent with expected criteria for
a 2.0 A structure, whereas both 1rfb and 2rig fell below acceptable criteria.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2 Stereoview of a portion of the electron-density map
superimposed on residues 36-55 for (a) the NCS-averaged
solvent-flattened MIR map, (b) the crystal form 1 2F[o] - F[c]
map (1  ,
2.9 Å) and (c) the crystal form 2 2F[o] - F[c] map (1 ,
2.9 Å) and (c) the crystal form 2 2F[o] - F[c] map (1  ,
2.0 Å). ,
2.0 Å).
|
 |
Figure 9.
Figure 9 (a) van der Waals surface representation of the
residues lining the buried cavity in bovine IFN- 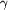 .
(b) Substitution of a low-energy conformation of Trp at residue
33 results in the elimination of this cavity. .
(b) Substitution of a low-energy conformation of Trp at residue
33 results in the elimination of this cavity.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2000,
56,
14-24)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.B.Trivella,
J.R.Ferreira-Júnior,
L.Dumoutier,
J.C.Renauld,
and
I.Polikarpov
(2010).
Structure and function of interleukin-22 and other members of the interleukin-10 family.
|
| |
Cell Mol Life Sci,
67,
2909-2935.
|
 |
|
|
|
|
 |
M.Yogavel,
S.Khan,
T.K.Bhatt,
and
A.Sharma
(2010).
Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
584-592.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Savan,
S.Ravichandran,
J.R.Collins,
M.Sakai,
and
H.A.Young
(2009).
Structural conservation of interferon gamma among vertebrates.
|
| |
Cytokine Growth Factor Rev,
20,
115-124.
|
 |
|
|
|
|
 |
C.D.Krause,
and
S.Pestka
(2005).
Evolution of the Class 2 cytokines and receptors, and discovery of new friends and relatives.
|
| |
Pharmacol Ther,
106,
299-346.
|
 |
|
|
|
|
 |
S.Pestka,
C.D.Krause,
D.Sarkar,
M.R.Walter,
Y.Shi,
and
P.B.Fisher
(2004).
Interleukin-10 and related cytokines and receptors.
|
| |
Annu Rev Immunol,
22,
929-979.
|
 |
|
|
|
|
 |
S.Pestka,
C.D.Krause,
and
M.R.Walter
(2004).
Interferons, interferon-like cytokines, and their receptors.
|
| |
Immunol Rev,
202,
8.
|
 |
|
|
|
|
 |
M.Randal,
and
A.A.Kossiakoff
(2001).
The structure and activity of a monomeric interferon-gamma:alpha-chain receptor signaling complex.
|
| |
Structure,
9,
155-163.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

