 |
PDBsum entry 1csh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase(oxo-acid)
|
PDB id
|
|

|

|
1csh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.3.1
- citrate (Si)-synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Citric acid cycle
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxaloacetate + acetyl-CoA + H2O = citrate + CoA + H+
|
 |
 |
 |
 |
 |
 oxaloacetate
oxaloacetate
|
+
|
 acetyl-CoA
acetyl-CoA
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
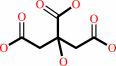 citrate
citrate
|
+
|
CoA
Bound ligand (Het Group name = )
matches with 90.38% similarity
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:7753-7759
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
A very short hydrogen bond provides only moderate stabilization of an enzyme-inhibitor complex of citrate synthase.
|
|
K.C.Usher,
S.J.Remington,
D.P.Martin,
D.G.Drueckhammer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Two extremely potent inhibitors of citrate synthase, carboxyl and primary amide
analogues of acetyl coenzyme A, have been synthesized. The ternary complexes of
these inhibitors with oxaloacetate and citrate synthase have been crystallized
and their structures analyzed at 1.70- and 1.65-A resolution, respectively. The
inhibitors have dissociation constants in the nanomolar range, with the carboxyl
analogue binding more tightly (Ki = 1.6 nM at pH 6.0) than the amide analogue
(28 nM), despite the unfavorable requirement for proton uptake by the former.
The carboxyl group forms a shorter hydrogen bond with the catalytic Asp 375
(distance < 2.4 A) than does the amide group (distance approximately 2.5 A).
Particularly with the carboxylate inhibitor, the very short hydrogen bond
distances measured suggest a low barrier or short strong hydrogen bond. However,
the binding constants differ by only a factor of 20 at pH 6.0, corresponding to
an increase in binding energy for the carboxyl analogue on the enzyme of about 2
kcal/mol more than the amide analogue, much less than has been proposed for
short strong hydrogen bonds based on gas phase measurements [> 20 kcal/mol
(Gerlt & Gassman, 1993a,b)]. The inhibitor complexes support proposals that
Asp 375 and His 274 work in concert to form an enolized form of acetyl-coenzyme
A as the first step in the reaction.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.C.Kurz,
C.Z.Constantine,
H.Jiang,
and
T.J.Kappock
(2009).
The partial substrate dethiaacetyl-coenzyme A mimics all critical carbon acid reactions in the condensation half-reaction catalyzed by Thermoplasma acidophilum citrate synthase.
|
| |
Biochemistry,
48,
7878-7891.
|
 |
|
|
|
|
 |
E.Chea,
and
D.R.Livesay
(2007).
How accurate and statistically robust are catalytic site predictions based on closeness centrality?
|
| |
BMC Bioinformatics,
8,
153.
|
 |
|
|
|
|
 |
M.W.van der Kamp,
F.Perruccio,
and
A.J.Mulholland
(2007).
Substrate polarization in enzyme catalysis: QM/MM analysis of the effect of oxaloacetate polarization on acetyl-CoA enolization in citrate synthase.
|
| |
Proteins,
69,
521-535.
|
 |
|
|
|
|
 |
C.C.Zhou,
K.D.Stewart,
and
M.K.Dhaon
(2005).
An intramolecular ionic hydrogen bond stabilizes a cis amide bond rotamer of a ring-opened rapamycin-degradation product.
|
| |
Magn Reson Chem,
43,
41-46.
|
 |
|
|
|
|
 |
M.Sugishima,
N.Tanimoto,
K.Soda,
N.Hamada,
F.Tokunaga,
and
K.Fukuyama
(2004).
Structure of photoactive yellow protein (PYP) E46Q mutant at 1.2 A resolution suggests how Glu46 controls the spectroscopic and kinetic characteristics of PYP.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2305-2309.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Anderson,
S.Crosson,
and
K.Moffat
(2004).
Short hydrogen bonds in photoactive yellow protein.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1008-1016.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Kumar,
and
R.Nussinov
(2004).
Different roles of electrostatics in heat and in cold: adaptation by citrate synthase.
|
| |
Chembiochem,
5,
280-290.
|
 |
|
|
|
|
 |
D.M.Anstrom,
K.Kallio,
and
S.J.Remington
(2003).
Structure of the Escherichia coli malate synthase G:pyruvate:acetyl-coenzyme A abortive ternary complex at 1.95 A resolution.
|
| |
Protein Sci,
12,
1822-1832.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.R.Howard,
J.A.Endrizzi,
and
S.J.Remington
(2000).
Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 A resolution: mechanistic implications.
|
| |
Biochemistry,
39,
3156-3168.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.M.Holtz,
B.Stec,
J.K.Myers,
S.M.Antonelli,
T.S.Widlanski,
and
E.R.Kantrowitz
(2000).
Alternate modes of binding in two crystal structures of alkaline phosphatase-inhibitor complexes.
|
| |
Protein Sci,
9,
907-915.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.C.Kurz,
G.Drysdale,
M.Riley,
M.A.Tomar,
J.Chen,
R.J.Russell,
and
M.J.Danson
(2000).
Kinetics and mechanism of the citrate synthase from the thermophilic archaeon Thermoplasma acidophilum.
|
| |
Biochemistry,
39,
2283-2296.
|
 |
|
|
|
|
 |
Z.Gu,
D.G.Drueckhammer,
L.Kurz,
K.Liu,
D.P.Martin,
and
A.McDermott
(1999).
Solid state NMR studies of hydrogen bonding in a citrate synthase inhibitor complex.
|
| |
Biochemistry,
38,
8022-8031.
|
 |
|
|
|
|
 |
L.C.Kurz,
T.Nakra,
R.Stein,
W.Plungkhen,
M.Riley,
F.Hsu,
and
G.R.Drysdale
(1998).
Effects of changes in three catalytic residues on the relative stabilities of some of the intermediates and transition states in the citrate synthase reaction.
|
| |
Biochemistry,
37,
9724-9737.
|
 |
|
|
|
|
 |
W.W.Cleland,
P.A.Frey,
and
J.A.Gerlt
(1998).
The low barrier hydrogen bond in enzymatic catalysis.
|
| |
J Biol Chem,
273,
25529-25532.
|
 |
|
|
|
|
 |
C.L.Perrin,
and
J.B.Nielson
(1997).
"Strong" hydrogen bonds in chemistry and biology.
|
| |
Annu Rev Phys Chem,
48,
511-544.
|
 |
|
|
|
|
 |
C.M.Wilmot,
J.M.Murray,
G.Alton,
M.R.Parsons,
M.A.Convery,
V.Blakeley,
A.S.Corner,
M.M.Palcic,
P.F.Knowles,
M.J.McPherson,
and
S.E.Phillips
(1997).
Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction.
|
| |
Biochemistry,
36,
1608-1620.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.A.Charlier,
C.Narasimhan,
and
H.M.Miziorko
(1997).
Inactivation of 3-hydroxy-3-methylglutaryl-CoA synthase and other Acyl-CoA-utilizing enzymes by 3-Oxobutylsulfoxyl-CoA.
|
| |
Biochemistry,
36,
1551-1558.
|
 |
|
|
|
|
 |
J.A.Gerlt,
M.M.Kreevoy,
W.Cleland,
and
P.A.Frey
(1997).
Understanding enzymic catalysis: the importance of short, strong hydrogen bonds.
|
| |
Chem Biol,
4,
259-267.
|
 |
|
|
|
|
 |
L.C.Kurz,
J.H.Roble,
T.Nakra,
G.R.Drysdale,
J.M.Buzan,
B.Schwartz,
and
D.G.Drueckhammer
(1997).
Ability of single-site mutants of citrate synthase to catalyze proton transfer from the methyl group of dethiaacetyl-coenzyme A, a non-thioester substrate analog.
|
| |
Biochemistry,
36,
3981-3990.
|
 |
|
|
|
|
 |
R.J.Russell,
J.M.Ferguson,
D.W.Hough,
M.J.Danson,
and
G.L.Taylor
(1997).
The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolution,.
|
| |
Biochemistry,
36,
9983-9994.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.T.Evans,
L.C.Kurz,
S.J.Remington,
and
P.A.Srere
(1996).
Active site mutants of pig citrate synthase: effects of mutations on the enzyme catalytic and structural properties.
|
| |
Biochemistry,
35,
10661-10672.
|
 |
|
|
|
|
 |
J.P.Guthrie
(1996).
Short strong hydrogen bonds: can they explain enzymic catalysis?
|
| |
Chem Biol,
3,
163-170.
|
 |
|
|
|
|
 |
O.Hur,
C.Leja,
and
M.F.Dunn
(1996).
Evidence of a low-barrier hydrogen bond in the tryptophan synthase catalytic mechanism.
|
| |
Biochemistry,
35,
7378-7386.
|
 |
|
|
|
|
 |
S.Bandyopadhyay,
C.Mukhopadhyay,
and
S.Roy
(1996).
Dimer-dimer interfaces of the lambda-repressor are different in liganded and free states.
|
| |
Biochemistry,
35,
5033-5040.
|
 |
|
|
|
|
 |
R.J.Russell,
D.W.Hough,
M.J.Danson,
and
G.L.Taylor
(1994).
The crystal structure of citrate synthase from the thermophilic archaeon, Thermoplasma acidophilum.
|
| |
Structure,
2,
1157-1167.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

