 |
PDBsum entry 1c3e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
New insights into inhibitor design from the crystal structure and nmr studies of e. Coli gar transformylate in complex with beta-gar and 10-formyl-5,8,10-trideazafolic acid.
|
|
Structure:
|
 |
Glycinamide ribonucleotide transformylase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: purn. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.227
|
R-free:
|
0.263
|
|
|
Authors:
|
 |
S.E.Greasley,M.M.Yamashita,H.Cai,S.J.Benkovic,D.L.Boger,I.A.Wilson
|
Key ref:

|
 |
S.E.Greasley
et al.
(1999).
New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry,
38,
16783-16793.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
27-Jul-99
|
Release date:
|
29-Dec-99
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P08179
(PUR3_ECOLI) -
Phosphoribosylglycinamide formyltransferase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
212 a.a.
209 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.2.2
- phosphoribosylglycinamide formyltransferase 1.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Purine Biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N1-(5-phospho-beta-D-ribosyl)glycinamide + (6R)-10- formyltetrahydrofolate = N2-formyl-N1-(5-phospho-beta-D- ribosyl)glycinamide + (6S)-5,6,7,8-tetrahydrofolate + H+
|
 |
 |
 |
 |
 |
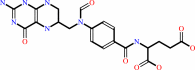 10-formyltetrahydrofolate
10-formyltetrahydrofolate
|
+
|
N(1)-(5-phospho-D-ribosyl)glycinamide
|
=
|
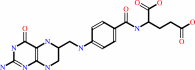 tetrahydrofolate
tetrahydrofolate
|
+
|
N(2)-formyl-N(1)-(5-phospho-D-ribosyl)glycinamide
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
38:16783-16793
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
|
|
S.E.Greasley,
M.M.Yamashita,
H.Cai,
S.J.Benkovic,
D.L.Boger,
I.A.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of Escherichia coli GAR Tfase at 2.1 A resolution in
complex with 10-formyl-5,8,10-trideazafolic acid (10-formyl-TDAF, K(i) = 260
nM), an inhibitor designed to form an enzyme-assembled multisubstrate adduct
with the substrate, beta-GAR, was studied to determine the exact nature of its
inhibitory properties. Rather than forming the expected covalent adduct, the
folate inhibitor binds as the hydrated aldehyde (gem-diol) in the enzyme active
site, in a manner that mimics the tetrahedral intermediate of the formyl
transfer reaction. In this hydrated form, the inhibitor not only provides
unexpected insights into the catalytic mechanism but also explains the 10-fold
difference in inhibitor potency between 10-formyl-TDAF and the corresponding
alcohol, and a further 10-fold difference for inhibitors that lack the alcohol.
The presence of the hydrated aldehyde was confirmed in solution by (13)C-(1)H
NMR spectroscopy of the ternary GAR Tfase-beta-GAR-10-formyl-TDAF complex using
the (13)C-labeled 10-formyl-TDAF. This insight into the behavior of the
inhibitor, which is analogous to protease or transaminase inhibitors, provides a
novel and previously unrecognized basis for the design of more potent inhibitors
of the folate-dependent formyl transfer enzymes of the purine biosynthetic
pathway and development of anti-neoplastic agents.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Pozharski
(2010).
Percentile-based spread: a more accurate way to compare crystallographic models.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
970-978.
|
 |
|
|
|
|
 |
J.K.DeMartino,
I.Hwang,
S.Connelly,
I.A.Wilson,
and
D.L.Boger
(2008).
Asymmetric synthesis of inhibitors of glycinamide ribonucleotide transformylase.
|
| |
J Med Chem,
51,
5441-5448.
|
 |
|
|
|
|
 |
Y.Zhang,
M.Morar,
and
S.E.Ealick
(2008).
Structural biology of the purine biosynthetic pathway.
|
| |
Cell Mol Life Sci,
65,
3699-3724.
|
 |
|
|
|
|
 |
T.C.Terwilliger,
R.W.Grosse-Kunstleve,
P.V.Afonine,
P.D.Adams,
N.W.Moriarty,
P.Zwart,
R.J.Read,
D.Turk,
and
L.W.Hung
(2007).
Interpretation of ensembles created by multiple iterative rebuilding of macromolecular models.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
597-610.
|
 |
|
|
|
|
 |
W.Manieri,
M.E.Moore,
M.B.Soellner,
P.Tsang,
and
C.A.Caperelli
(2007).
Human glycinamide ribonucleotide transformylase: active site mutants as mechanistic probes.
|
| |
Biochemistry,
46,
156-163.
|
 |
|
|
|
|
 |
P.Kursula,
H.Schüler,
S.Flodin,
P.Nilsson-Ehle,
D.J.Ogg,
P.Savitsky,
P.Nordlund,
and
P.Stenmark
(2006).
Structures of the hydrolase domain of human 10-formyltetrahydrofolate dehydrogenase and its complex with a substrate analogue.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1294-1299.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.N.Reuland,
A.P.Vlasov,
and
S.A.Krupenko
(2006).
Modular organization of FDH: Exploring the basis of hydrolase catalysis.
|
| |
Protein Sci,
15,
1076-1084.
|
 |
|
|
|
|
 |
L.Xu,
C.Li,
A.J.Olson,
and
I.A.Wilson
(2004).
Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening.
|
| |
J Biol Chem,
279,
50555-50565.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Morikis,
A.H.Elcock,
P.A.Jennings,
and
J.A.McCammon
(2001).
Native-state conformational dynamics of GART: a regulatory pH-dependent coil-helix transition examined by electrostatic calculations.
|
| |
Protein Sci,
10,
2363-2378.
|
 |
|
|
|
|
 |
D.Morikis,
A.H.Elcock,
P.A.Jennings,
and
J.A.McCammon
(2001).
Proton transfer dynamics of GART: the pH-dependent catalytic mechanism examined by electrostatic calculations.
|
| |
Protein Sci,
10,
2379-2392.
|
 |
|
|
|
|
 |
V.M.Reyes,
S.E.Greasley,
E.A.Stura,
G.P.Beardsley,
and
I.A.Wilson
(2000).
Crystallization and preliminary crystallographic investigations of avian 5-aminoimidazole-4-carboxamide ribonucleotide transformylase-inosine monophosphate cyclohydrolase expressed in Escherichia coli.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
1051-1054.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

