 |
PDBsum entry 1bvh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.2
- acid phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a phosphate monoester + H2O = an alcohol + phosphate
|
 |
 |
 |
 |
 |
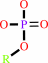 phosphate monoester
phosphate monoester
|
+
|
H2O
|
=
|
 alcohol
alcohol
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.3.48
- protein-tyrosine-phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-phospho-L-tyrosyl-[protein] + H2O = L-tyrosyl-[protein] + phosphate
|
 |
 |
 |
 |
 |
O-phospho-L-tyrosyl-[protein]
|
+
|
H2O
|
=
|
L-tyrosyl-[protein]
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:11087-11096
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Solution structure of a low molecular weight protein tyrosine phosphatase.
|
|
T.M.Logan,
M.M.Zhou,
D.G.Nettesheim,
R.P.Meadows,
R.L.Van Etten,
S.W.Fesik.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Protein tyrosine phosphatases (PTPs) are important enzymes involved in signal
transduction, cell cycle regulation, and the control of differentiation. Despite
the importance of this class of enzymes in the control of critical cell
processes, very little structural information is available for this family of
proteins. In this paper, we present the first solution structure of a protein
tyrosine phosphatase. This protein is a low molecular weight cytosolic PTP that
was initially isolated from bovine heart. The structure that was determined from
1747 NMR-derived restraints consists of a central four-stranded parallel
beta-sheet surrounded by four alpha-helices and a short 3(10) helix. The
phosphate binding site, identified by chemical shift changes upon the addition
of the competitive inhibitors phosphate and vanadate, is in a loop region
connecting the C-terminal end of the first beta-strand with the first
alpha-helix. Residues in the second, fourth, and fifth alpha-helices and in some
of the loop regions connecting the elements of regular secondary structure also
contribute to the binding site. The structure determined here is consistent with
previous mutagenesis and chemical modification studies conducted on this protein.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Cunha,
M.Kuti,
D.F.Bishop,
M.Mezei,
L.Zeng,
M.M.Zhou,
and
R.J.Desnick
(2008).
Human uroporphyrinogen III synthase: NMR-based mapping of the active site.
|
| |
Proteins,
71,
855-873.
|
 |
|
|
|
|
 |
D.Tolkatchev,
R.Shaykhutdinov,
P.Xu,
J.Plamondon,
D.C.Watson,
N.M.Young,
and
F.Ni
(2006).
Three-dimensional structure and ligand interactions of the low molecular weight protein tyrosine phosphatase from Campylobacter jejuni.
|
| |
Protein Sci,
15,
2381-2394.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Lescop,
Y.Hu,
H.Xu,
W.Hu,
J.Chen,
B.Xia,
and
C.Jin
(2006).
The solution structure of Escherichia coli Wzb reveals a novel substrate recognition mechanism of prokaryotic low molecular weight protein-tyrosine phosphatases.
|
| |
J Biol Chem,
281,
19570-19577.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Xu,
B.Xia,
and
C.Jin
(2006).
Solution structure of a low-molecular-weight protein tyrosine phosphatase from Bacillus subtilis.
|
| |
J Bacteriol,
188,
1509-1517.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.L.Gustafson,
C.V.Stauffacher,
K.Hallenga,
and
R.L.Van Etten
(2005).
Solution structure of the low-molecular-weight protein tyrosine phosphatase from Tritrichomonas foetus reveals a flexible phosphate binding loop.
|
| |
Protein Sci,
14,
2515-2525.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.M.Legler,
M.Cai,
A.Peterkofsky,
and
G.M.Clore
(2004).
Three-dimensional solution structure of the cytoplasmic B domain of the mannitol transporter IImannitol of the Escherichia coli phosphotransferase system.
|
| |
J Biol Chem,
279,
39115-39121.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Lah,
J.Lah,
I.Zegers,
L.Wyns,
and
J.Messens
(2003).
Specific potassium binding stabilizes pI258 arsenate reductase from Staphylococcus aureus.
|
| |
J Biol Chem,
278,
24673-24679.
|
 |
|
|
|
|
 |
I.Zegers,
J.C.Martins,
R.Willem,
L.Wyns,
and
J.Messens
(2001).
Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty.
|
| |
Nat Struct Biol,
8,
843-847.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Wang,
L.Tabernero,
M.Zhang,
E.Harms,
R.L.Van Etten,
and
C.V.Stauffacher
(2000).
Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
|
| |
Biochemistry,
39,
1903-1914.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Kolmodin,
P.Nordlund,
and
J.Aqvist
(1999).
Mechanism of substrate dephosphorylation in low Mr protein tyrosine phosphatase.
|
| |
Proteins,
36,
370-379.
|
 |
|
|
|
|
 |
M.Zhou,
and
R.L.Van Etten
(1999).
Structural basis of the tight binding of pyridoxal 5'-phosphate to a low molecular weight protein tyrosine phosphatase.
|
| |
Biochemistry,
38,
2636-2646.
|
 |
|
|
|
|
 |
M.Zhang,
C.V.Stauffacher,
D.Lin,
and
R.L.Van Etten
(1998).
Crystal structure of a human low molecular weight phosphotyrosyl phosphatase. Implications for substrate specificity.
|
| |
J Biol Chem,
273,
21714-21720.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Ramponi,
and
M.Stefani
(1997).
Structural, catalytic, and functional properties of low M(r), phosphotyrosine protein phosphatases. Evidence of a long evolutionary history.
|
| |
Int J Biochem Cell Biol,
29,
279-292.
|
 |
|
|
|
|
 |
M.J.Pregel,
and
A.C.Storer
(1997).
Active site titration of the tyrosine phosphatases SHP-1 and PTP1B using aromatic disulfides. Reaction with the essential cysteine residue in the active site.
|
| |
J Biol Chem,
272,
23552-23558.
|
 |
|
|
|
|
 |
M.Zhang,
M.Zhou,
R.L.Van Etten,
and
C.V.Stauffacher
(1997).
Crystal structure of bovine low molecular weight phosphotyrosyl phosphatase complexed with the transition state analog vanadate.
|
| |
Biochemistry,
36,
15-23.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.A.Tishmack,
D.Bashford,
E.Harms,
and
R.L.Van Etten
(1997).
Use of 1H NMR spectroscopy and computer simulations To analyze histidine pKa changes in a protein tyrosine phosphatase: experimental and theoretical determination of electrostatic properties in a small protein.
|
| |
Biochemistry,
36,
11984-11994.
|
 |
|
|
|
|
 |
B.Evans,
P.A.Tishmack,
C.Pokalsky,
M.Zhang,
and
R.L.Van Etten
(1996).
Site-directed mutagenesis, kinetic, and spectroscopic studies of the P-loop residues in a low molecular weight protein tyrosine phosphatase.
|
| |
Biochemistry,
35,
13609-13617.
|
 |
|
|
|
|
 |
J.W.Eckstein,
P.Beer-Romero,
and
I.Berdo
(1996).
Identification of an essential acidic residue in Cdc25 protein phosphatase and a general three-dimensional model for a core region in protein phosphatases.
|
| |
Protein Sci,
5,
5.
|
 |
|
|
|
|
 |
X.Xu,
and
S.P.Burke
(1996).
Roles of active site residues and the NH2-terminal domain in the catalysis and substrate binding of human Cdc25.
|
| |
J Biol Chem,
271,
5118-5124.
|
 |
|
|
|
|
 |
C.Pokalsky,
P.Wick,
E.Harms,
F.E.Lytle,
and
R.L.Van Etten
(1995).
Fluorescence resolution of the intrinsic tryptophan residues of bovine protein tyrosyl phosphatase.
|
| |
J Biol Chem,
270,
3809-3815.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

