 |
PDBsum entry 1bsv

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1bsv
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.271
- GDP-L-fucose synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
GDP-L-Fucose and GDP-mannose Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
GDP-beta-L-fucose + NADP+ = GDP-4-dehydro-alpha-D-rhamnose + NADPH + H+
|
 |
 |
 |
 |
 |
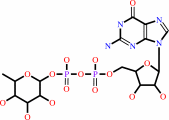 GDP-beta-L-fucose
GDP-beta-L-fucose
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
GDP-4-dehydro-alpha-D-rhamnose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
6:1601-1612
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site.
|
|
W.S.Somers,
M.L.Stahl,
F.X.Sullivan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Background:. In all species examined, GDP-fucose is synthesized from GDP-mannose
in a three-step reaction catalyzed by two enzymes, GDP-mannose 4,6 dehydratase
and a dual function 3, 5-epimerase-4-reductase named GDP-fucose synthetase. In
this latter aspect fucose biosynthesis differs from that of other deoxy and
dideoxy sugars, in which the epimerase and reductase activities are present as
separate enzymes. Defects in GDP-fucose biosynthesis have been shown to affect
nodulation in bacteria, stem development in plants, and are associated with the
immune defect leukocyte adhesion deficiency type II in humans. Results:. We have
determined the structure of GDP-fucose synthetase from Escherichia coli at 2.2 A
resolution. The structure of GDP-fucose synthetase is closely related to that of
UDP-galactose 4-epimerase and more distantly to other members of the short-chain
dehydrogenase/reductase family. We have also determined the structures of the
binary complexes of GDP-fucose synthetase with its substrate NADPH and its
product NADP+. The nicotinamide cofactors bind in the syn and anti
conformations, respectively. Conclusions:. GDP-fucose synthetase binds its
substrate, NADPH, in the proper orientation (syn) for transferring the 4-pro-S
hydride of the nicotinamide. We have observed a single binding site in
GDP-fucose synthetase for the second substrate, GDP-4-keto,6-deoxy-mannose. This
implies that both the epimerization and reduction reactions occur at the same
site in the enzyme. As is the case for all members of the short-chain family of
dehydrogenase/reductases, GDP-fucose synthetase retains the Ser-Tyr-Lys
catalytic triad. We propose that this catalytic triad functions in a
mechanistically equivalent manner in both the epimerization and reduction
reactions. Additionally, the X-ray structure has allowed us to identify other
residues that are potentially required for substrate binding and catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. A ball-and-stick representation of the
GDP-4-keto-6 deoxy-mannose binding model. The proposed
binding-site residues are shown with dark bonds and the
substrate/NADPH nicotinamide ring is shown with light-grey bonds.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1998,
6,
1601-1612)
copyright 1998.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.H.Serres,
A.R.Kerr,
T.J.McCormack,
and
M.Riley
(2009).
Evolution by leaps: gene duplication in bacteria.
|
| |
Biol Direct,
4,
46.
|
 |
|
|
|
|
 |
C.J.Thibodeaux,
C.E.Melançon,
and
H.W.Liu
(2008).
Natural-product sugar biosynthesis and enzymatic glycodiversification.
|
| |
Angew Chem Int Ed Engl,
47,
9814-9859.
|
 |
|
|
|
|
 |
C.Dong,
L.L.Major,
V.Srikannathasan,
J.C.Errey,
M.F.Giraud,
J.S.Lam,
M.Graninger,
P.Messner,
M.R.McNeil,
R.A.Field,
C.Whitfield,
and
J.H.Naismith
(2007).
RmlC, a C3' and C5' carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation.
|
| |
J Mol Biol,
365,
146-159.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.H.Ehrensberger,
R.A.Elling,
and
D.K.Wilson
(2006).
Structure-guided engineering of xylitol dehydrogenase cosubstrate specificity.
|
| |
Structure,
14,
567-575.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.A.Webb,
A.M.Mulichak,
J.S.Lam,
H.L.Rocchetta,
and
R.M.Garavito
(2004).
Crystal structure of a tetrameric GDP-D-mannose 4,6-dehydratase from a bacterial GDP-D-rhamnose biosynthetic pathway.
|
| |
Protein Sci,
13,
529-539.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Brandt,
M.A.Dessoy,
M.Fulhorst,
W.Gao,
M.H.Zenk,
and
L.A.Wessjohann
(2004).
A proposed mechanism for the reductive ring opening of the cyclodiphosphate MEcPP, a crucial transformation in the new DXP/MEP pathway to isoprenoids based on modeling studies and feeding experiments.
|
| |
Chembiochem,
5,
311-323.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.A.Wolucka,
and
M.Van Montagu
(2003).
GDP-mannose 3',5'-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants.
|
| |
J Biol Chem,
278,
47483-47490.
|
 |
|
|
|
|
 |
e.l.-.S.E.Habib,
J.N.Scarsdale,
and
K.A.Reynolds
(2003).
Biosynthetic origin of hygromycin A.
|
| |
Antimicrob Agents Chemother,
47,
2065-2071.
|
 |
|
|
|
|
 |
C.Creuzenet,
R.V.Urbanic,
and
J.S.Lam
(2002).
Structure-function studies of two novel UDP-GlcNAc C6 dehydratases/C4 reductases. Variation from the SYK dogma.
|
| |
J Biol Chem,
277,
26769-26778.
|
 |
|
|
|
|
 |
B.A.Wolucka,
G.Persiau,
J.Van Doorsselaere,
M.W.Davey,
H.Demol,
J.Vandekerckhove,
M.Van Montagu,
M.Zabeau,
and
W.Boerjan
(2001).
Partial purification and identification of GDP-mannose 3",5"-epimerase of Arabidopsis thaliana, a key enzyme of the plant vitamin C pathway.
|
| |
Proc Natl Acad Sci U S A,
98,
14843-14848.
|
 |
|
|
|
|
 |
E.Maser,
G.Xiong,
C.Grimm,
R.Ficner,
and
K.Reuter
(2001).
3alpha-Hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni: biological significance, three-dimensional structure and gene regulation.
|
| |
Chem Biol Interact,
130,
707-722.
|
 |
|
|
|
|
 |
A.M.Deacon,
Y.S.Ni,
W.G.Coleman,
and
S.E.Ealick
(2000).
The crystal structure of ADP-L-glycero-D-mannoheptose 6-epimerase: catalysis with a twist.
|
| |
Structure,
8,
453-462.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Benach,
S.Atrian,
J.Fibla,
R.Gonzàlez-Duarte,
and
R.Ladenstein
(2000).
Structure-function relationships in Drosophila melanogaster alcohol dehydrogenase allozymes ADH(S), ADH(F) and ADH(UF), and distantly related forms.
|
| |
Eur J Biochem,
267,
3613-3622.
|
 |
|
|
|
|
 |
J.R.Somoza,
S.Menon,
H.Schmidt,
D.Joseph-McCarthy,
A.Dessen,
M.L.Stahl,
W.S.Somers,
and
F.X.Sullivan
(2000).
Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme's catalytic mechanism and regulation by GDP-fucose.
|
| |
Structure,
8,
123-135.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.F.Giraud,
and
J.H.Naismith
(2000).
The rhamnose pathway.
|
| |
Curr Opin Struct Biol,
10,
687-696.
|
 |
|
|
|
|
 |
M.Riley,
and
M.H.Serres
(2000).
Interim report on genomics of Escherichia coli.
|
| |
Annu Rev Microbiol,
54,
341-411.
|
 |
|
|
|
|
 |
A.M.Mulichak,
M.J.Theisen,
B.Essigmann,
C.Benning,
and
R.M.Garavito
(1999).
Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose.
|
| |
Proc Natl Acad Sci U S A,
96,
13097-13102.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Graninger,
B.Nidetzky,
D.E.Heinrichs,
C.Whitfield,
and
P.Messner
(1999).
Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-L-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2.
|
| |
J Biol Chem,
274,
25069-25077.
|
 |
|
|
|
|
 |
S.Menon,
M.Stahl,
R.Kumar,
G.Y.Xu,
and
F.Sullivan
(1999).
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from Escherichia coli.
|
| |
J Biol Chem,
274,
26743-26750.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

