 |
PDBsum entry 1bpn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase(alpha-aminoacylpeptide)
|
PDB id
|
|

|

|
1bpn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.4.11.1
- leucyl aminopeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Release of an N-terminal amino acid, Xaa-|-Xbb-, in which Xaa is preferably Leu, but may be other amino acids including Pro although not Arg or Lys, and Xbb may be Pro.
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.4.11.5
- prolyl aminopeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Release of a N-terminal proline from a peptide.
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.4.13.23
- cysteinylglycine-S-conjugate dipeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an S-substituted L-cysteinylglycine + H2O = an S-substituted L-cysteine + glycine
|
 |
 |
 |
 |
 |
S-substituted L-cysteinylglycine
|
+
|
H2O
|
=
|
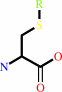 S-substituted L-cysteine
S-substituted L-cysteine
|
+
|
 glycine
glycine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
90:5006-5010
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Differentiation and identification of the two catalytic metal binding sites in bovine lens leucine aminopeptidase by x-ray crystallography.
|
|
H.Kim,
W.N.Lipscomb.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The tightly binding and readily exchanging metal binding sites in the active
site of bovine lens leucine aminopeptidase (blLAP; EC 3.4.11.1) have been
differentiated and identified by x-ray crystallography. In native blLAP,Zn2+
occupies both binding sites. In solution, site 1 readily exchanges Zn2+ for
other divalent cations, including Mg2+. The Zn2+ in site 2 is unavailable for
metal exchange under conditions which allow exchange at site 1. The Zn2+/Mg2+
metal hybrid of blLAP (Mg-blLAP) was prepared in solution and crystallized.
X-ray diffraction data to 2.9-A resolution were collected at -150 degrees C from
single crystals of Mg-blLAP and native blLAP. Comparisons of omit maps
calculated from the Mg-blLAP data with analogous maps calculated from the native
blLAP data show electron density in one of the metal binding sites in Mg-blLAP
which is much weaker than the electron density in the other binding site. Since
there are fewer electrons associated with Mg2+ than with Zn2+, the difference in
electron density between the two metal binding sites is consistent with
occupancy of the weaker electron density site by Mg2+ and identifies this metal
binding site as site 1, defined as the readily exchanging site. The present
identification of the metal binding sites reverses the previous presumptive
assignment of the metal binding sites which was based on the structure of native
blLAP [Burley, S. K., David, P. R., Sweet, R. M., Taylor, A. & Lipscomb, W.
N. (1992) J. Mol. Biol. 224, 113-140]. According to the residue-numbering
convention of native blLAP, the new assignment of the metal binding sites
identifies the readily exchanging site 1 with Zn-488, which is within
interaction distance of one side-chain carboxylate oxygen from each of Asp-255,
Asp-332, and Glu-334 and the main-chain carbonyl oxygen of Asp-332. The more
tightly binding site 2 is identified with Zn-489, which is within interaction
distance of one side-chain carboxylate oxygen from each of Asp-255, Asp-273, and
Glu-334 and the side-chain amine nitrogen of Lys-250.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.H.Chen,
M.J.Cao,
J.Z.Huang,
and
G.P.Wu
(2011).
Identification of a puromycin-sensitive aminopeptidase from zebrafish (Danio rerio).
|
| |
Comp Biochem Physiol B Biochem Mol Biol,
159,
10-17.
|
 |
|
|
|
|
 |
L.G.Zhou,
B.X.Liu,
L.C.Sun,
K.Hara,
W.J.Su,
and
M.J.Cao
(2010).
Identification of an aminopeptidase from the skeletal muscle of grass carp (Ctenopharyngodon idellus).
|
| |
Fish Physiol Biochem,
36,
953-962.
|
 |
|
|
|
|
 |
M.C.Chi,
H.B.Huang,
J.S.Liu,
W.C.Wang,
W.C.Liang,
and
L.L.Lin
(2006).
Residues threonine 346 and leucine 352 are critical for the proper function of Bacillus kaustophilus leucine aminopeptidase.
|
| |
FEMS Microbiol Lett,
260,
156-161.
|
 |
|
|
|
|
 |
Q.Z.Ye,
S.X.Xie,
Z.Q.Ma,
M.Huang,
and
R.P.Hanzlik
(2006).
Structural basis of catalysis by monometalated methionine aminopeptidase.
|
| |
Proc Natl Acad Sci U S A,
103,
9470-9475.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.B.Huang,
M.C.Chi,
W.H.Hsu,
W.C.Liang,
and
L.L.Lin
(2005).
Construction and one-step purification of Bacillus kaustophilus leucine aminopeptidase fused to the starch-binding domain of Bacillus sp. strain TS-23 alpha-amylase.
|
| |
Bioprocess Biosyst Eng,
27,
389-398.
|
 |
|
|
|
|
 |
S.V.Story,
C.Shah,
F.E.Jenney,
and
M.W.Adams
(2005).
Characterization of a novel zinc-containing, lysine-specific aminopeptidase from the hyperthermophilic archaeon Pyrococcus furiosus.
|
| |
J Bacteriol,
187,
2077-2083.
|
 |
|
|
|
|
 |
M.C.Chi,
W.M.Chou,
W.H.Hsu,
and
L.L.Lin
(2004).
Identification of amino acid residues essential for the catalytic reaction of Bacillus kaustophilus leucine aminopeptidase.
|
| |
Biosci Biotechnol Biochem,
68,
1794-1797.
|
 |
|
|
|
|
 |
D.H.Broder,
and
C.G.Miller
(2003).
DapE can function as an aspartyl peptidase in the presence of Mn2+.
|
| |
J Bacteriol,
185,
4748-4754.
|
 |
|
|
|
|
 |
S.Erhardt,
and
J.Weston
(2002).
Development of a working model of the active site in bovine lens leucine aminopeptidase: a density functional investigation.
|
| |
Chembiochem,
3,
101-104.
|
 |
|
|
|
|
 |
Y.Q.Gu,
F.M.Holzer,
and
L.L.Walling
(1999).
Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of tomato.
|
| |
Eur J Biochem,
263,
726-735.
|
 |
|
|
|
|
 |
B.Bennett,
and
R.C.Holz
(1997).
Spectroscopically distinct cobalt(II) sites in heterodimetallic forms of the aminopeptidase from Aeromonas proteolytica: characterization of substrate binding.
|
| |
Biochemistry,
36,
9837-9846.
|
 |
|
|
|
|
 |
G.Chen,
T.Edwards,
V.M.D'souza,
and
R.C.Holz
(1997).
Mechanistic studies on the aminopeptidase from Aeromonas proteolytica: a two-metal ion mechanism for peptide hydrolysis.
|
| |
Biochemistry,
36,
4278-4286.
|
 |
|
|
|
|
 |
B.Chevrier,
C.Schalk,
H.D'Orchymont,
J.M.Rondeau,
D.Moras,
and
C.Tarnus
(1994).
Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family.
|
| |
Structure,
2,
283-291.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

