 |
PDBsum entry 1bf3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1bf3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.13.2
- 4-hydroxybenzoate 3-monooxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxybenzoate + NADPH + O2 + H+ = 3,4-dihydroxybenzoate + NADP+ + H2O
|
 |
 |
 |
 |
 |
4-hydroxybenzoate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADPH
NADPH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
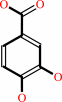 3,4-dihydroxybenzoate
3,4-dihydroxybenzoate
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Eur J Biochem
253:194-201
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Lys42 and Ser42 variants of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens reveal that Arg42 is essential for NADPH binding.
|
|
M.H.Eppink,
H.A.Schreuder,
W.J.van Berkel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The conserved Arg42 of the flavoprotein p-hydroxybenzoate hydroxylase is located
at the entrance of the active site in a loop between helix H2 and sheet E1 of
the FAD-binding domain. Replacement of Arg42 by Lys or Ser decreases the
turnover rate of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens by
more than two orders of magnitude. Rapid reaction kinetics show that the low
activity of the Arg42 variants results from impaired binding of NADPH. In
contrast to an earlier conclusion drawn for p-hydroxybenzoate hydroxylase from
Acinetobacter calcoaceticus, substitution of Arg42 with Ser42 in the enzyme from
P. fluorescens hardly disturbs the binding of FAD. Crystals of
[Lys42]p-hydroxybenzoate hydroxylase complexed with 4-hydroxybenzoate diffract
to 0.22-nm resolution. The structure of the Lys42 variant is virtually
indistinguishable from the native enzyme with the flavin ring occupying the
interior position within the active site. Lys42 in the mutant structure
interacts indirectly via a solvent molecule with the 3-OH of the adenosine
ribose moiety of FAD. Substrate perturbation difference spectra suggest that the
Arg42 replacements influence the solvent accessibility of the flavin ring in the
oxidized enzyme. In spite of this, the Arg42 variants fully couple enzyme
reduction to substrate hydroxylation. Sequence-comparison studies suggest that
Arg42 is involved in binding of the 2'-phosphoadenosine moiety of NADPH.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Kasai,
T.Fujinami,
T.Abe,
K.Mase,
Y.Katayama,
M.Fukuda,
and
E.Masai
(2009).
Uncovering the protocatechuate 2,3-cleavage pathway genes.
|
| |
J Bacteriol,
191,
6758-6768.
|
 |
|
|
|
|
 |
Y.Huang,
K.X.Zhao,
X.H.Shen,
C.Y.Jiang,
and
S.J.Liu
(2008).
Genetic and biochemical characterization of a 4-hydroxybenzoate hydroxylase from Corynebacterium glutamicum.
|
| |
Appl Microbiol Biotechnol,
78,
75-83.
|
 |
|
|
|
|
 |
C.Siebold,
N.Berrow,
T.S.Walter,
K.Harlos,
R.J.Owens,
D.I.Stuart,
J.R.Terman,
A.L.Kolodkin,
R.J.Pasterkamp,
and
E.Y.Jones
(2005).
High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule.
|
| |
Proc Natl Acad Sci U S A,
102,
16836-16841.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Wang,
M.Ortiz-Maldonado,
B.Entsch,
V.Massey,
D.Ballou,
and
D.L.Gatti
(2002).
Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
|
| |
Proc Natl Acad Sci U S A,
99,
608-613.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Dym,
and
D.Eisenberg
(2001).
Sequence-structure analysis of FAD-containing proteins.
|
| |
Protein Sci,
10,
1712-1728.
|
 |
|
|
|
|
 |
M.H.Eppink,
H.A.Schreuder,
and
W.J.van Berkel
(1998).
Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
|
| |
J Biol Chem,
273,
21031-21039.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

