 |
PDBsum entry 1bdb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1bdb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase from pseudomonas sp. Lb400
|
|
Structure:
|
 |
Cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase. Chain: a
|
|
Source:
|
 |
Pseudomonas sp.. Organism_taxid: 306. Strain: lb400. Variant: plebd4. Cellular_location: cytoplasm
|
|
Biol. unit:
|
 |
 Tetramer (from PDB file)
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.179
|
R-free:
|
0.230
|
|
|
Authors:
|
 |
M.Huelsmeyer,H.-J.Hecht,K.Niefind,B.Hofer,K.N.Timmis,D.Schomburg
|
Key ref:

|
 |
M.Hülsmeyer
et al.
(1998).
Crystal structure of cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase from a PCB degrader at 2.0 A resolution.
Protein Sci,
7,
1286-1293.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
10-May-97
|
Release date:
|
12-Nov-97
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P47227
(BPHB_PARXL) -
Cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase from Paraburkholderia xenovorans (strain LB400)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
277 a.a.
267 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.1.56
- cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-phenylcyclohexa-3,5-diene-1,2-diol + NAD+ = biphenyl-2,3-diol + NADH + H+
|
 |
 |
 |
 |
 |
(2R,3S)-3-phenylcyclohexa-3,5-diene-1,2-diol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
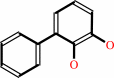 biphenyl-2,3-diol
biphenyl-2,3-diol
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
7:1286-1293
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase from a PCB degrader at 2.0 A resolution.
|
|
M.Hülsmeyer,
H.J.Hecht,
K.Niefind,
B.Hofer,
L.D.Eltis,
K.N.Timmis,
D.Schomburg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB) is involved in the aerobic
biodegradation of polychlorinated biphenyls (PCBs). The crystal structure of the
NAD+-enzyme complex was determined by molecular replacement and refined to an
R-value of 17.9% at 2.0 A. As a member of the short-chain alcohol
dehydrogenase/reductase (SDR) family, the overall protein fold and positioning
of the catalytic triad in BphB are very similar to those observed in other SDR
enzymes, although small differences occur in the cofactor binding site. Modeling
studies indicate that the substrate is bound in a deep hydrophobic cleft close
to the nicotinamide moiety of the NAD+ cofactor. These studies further suggest
that Asn143 is a key determinant of substrate specificity. A two-step reaction
mechanism is proposed for cis-dihydrodiol dehydrogenases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Fig. 5. Stereoscopic view f the active site withelectrondensitycontoured at Thedockedsubstrate BPDD (ontheright)hasbeen
incorporated to thepicture.
|
 |
Figure 7.
Fig. 7. Proposed reaction mechanism.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the Protein Society:
Protein Sci
(1998,
7,
1286-1293)
copyright 1998.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Xu,
J.J.Xu,
L.Y.Jia,
W.B.Liu,
and
X.Jian
(2011).
Congener Selectivity During Polychlorinated Biphenyls Degradation by Enterobacter sp. LY402.
|
| |
Curr Microbiol,
62,
784-789.
|
 |
|
|
|
|
 |
K.E.van Straaten,
H.Zheng,
D.R.Palmer,
and
D.A.Sanders
(2010).
Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
|
| |
Biochem J,
432,
237-247.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Vega-López,
F.A.Jiménez-Orozco,
L.A.Jiménez-Zamudio,
E.García-Latorre,
and
M.L.Domínguez-López
(2009).
Phase I enzyme induction in Girardinichthys viviparus, an endangered goodeid fish, exposed to water from native localities enriched with polychlorinated biphenyls.
|
| |
Arch Environ Contam Toxicol,
57,
561-570.
|
 |
|
|
|
|
 |
L.F.Pacios,
V.M.Campos,
I.Merino,
and
L.Gómez
(2009).
Structures and thermodynamics of biphenyl dihydrodiol stereoisomers and their metabolites in the enzymatic degradation of arene xenobiotics.
|
| |
J Comput Chem,
30,
2420-2432.
|
 |
|
|
|
|
 |
A.Vega-López,
L.Martínez-Tabche,
and
M.G.Martínez
(2007).
Toxic effects of waterborne polychlorinated biphenyls and sex differences in an endangered goodeid fish (Girardinichthys viviparus).
|
| |
Environ Int,
33,
540-545.
|
 |
|
|
|
|
 |
H.Cho,
L.Huang,
A.Hamza,
D.Gao,
C.G.Zhan,
and
H.H.Tai
(2006).
Role of glutamine 148 of human 15-hydroxyprostaglandin dehydrogenase in catalytic oxidation of prostaglandin E2.
|
| |
Bioorg Med Chem,
14,
6486-6491.
|
 |
|
|
|
|
 |
Y.Jouanneau,
and
C.Meyer
(2006).
Purification and characterization of an arene cis-dihydrodiol dehydrogenase endowed with broad substrate specificity toward polycyclic aromatic hydrocarbon dihydrodiols.
|
| |
Appl Environ Microbiol,
72,
4726-4734.
|
 |
|
|
|
|
 |
D.H.Pieper
(2005).
Aerobic degradation of polychlorinated biphenyls.
|
| |
Appl Microbiol Biotechnol,
67,
170-191.
|
 |
|
|
|
|
 |
A.Berchanski,
B.Shapira,
and
M.Eisenstein
(2004).
Hydrophobic complementarity in protein-protein docking.
|
| |
Proteins,
56,
130-142.
|
 |
|
|
|
|
 |
A.Berchanski,
and
M.Eisenstein
(2003).
Construction of molecular assemblies via docking: modeling of tetramers with D2 symmetry.
|
| |
Proteins,
53,
817-829.
|
 |
|
|
|
|
 |
W.L.Duax,
V.Pletnev,
A.Addlagatta,
J.Bruenn,
and
C.M.Weeks
(2003).
Rational proteomics I. Fingerprint identification and cofactor specificity in the short-chain oxidoreductase (SCOR) enzyme family.
|
| |
Proteins,
53,
931-943.
|
 |
|
|
|
|
 |
E.Díaz,
A.Ferrández,
M.A.Prieto,
and
J.L.García
(2001).
Biodegradation of aromatic compounds by Escherichia coli.
|
| |
Microbiol Mol Biol Rev,
65,
523.
|
 |
|
|
|
|
 |
E.T.Johnson,
S.Ryu,
H.Yi,
B.Shin,
H.Cheong,
and
G.Choi
(2001).
Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase.
|
| |
Plant J,
25,
325-333.
|
 |
|
|
|
|
 |
M.Otagiri,
G.Kurisu,
S.Swaminathan,
S.Ui,
S.Yoneda,
M.Ohkuma,
T.Kudo,
and
M.Kusunoki
(2001).
Crystallization and preliminary X-ray studies of meso-2,3-butanediol dehydrogenase from Klebsiella pneumoniae IAM1063.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
857-859.
|
 |
|
|
|
|
 |
T.Lanisnik Rizner,
J.Stojan,
and
J.Adamski
(2001).
17beta-hydroxysteroid dehydrogenase from the fungus Cochliobolus lunatus: structural and functional aspects.
|
| |
Chem Biol Interact,
130,
793-803.
|
 |
|
|
|
|
 |
J.Benach,
S.Atrian,
J.Fibla,
R.Gonzàlez-Duarte,
and
R.Ladenstein
(2000).
Structure-function relationships in Drosophila melanogaster alcohol dehydrogenase allozymes ADH(S), ADH(F) and ADH(UF), and distantly related forms.
|
| |
Eur J Biochem,
267,
3613-3622.
|
 |
|
|
|
|
 |
M.Vedadi,
D.Barriault,
M.Sylvestre,
and
J.Powlowski
(2000).
Active site residues of cis-2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase from Comamonas testosteroni strain B-356.
|
| |
Biochemistry,
39,
5028-5034.
|
 |
|
|
|
|
 |
R.E.Parales,
S.M.Resnick,
C.L.Yu,
D.R.Boyd,
N.D.Sharma,
and
D.T.Gibson
(2000).
Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the alpha subunit.
|
| |
J Bacteriol,
182,
5495-5504.
|
 |
|
|
|
|
 |
F.Brühlmann,
and
W.Chen
(1999).
Transformation of polychlorinated biphenyls by a novel BphA variant through the meta-cleavage pathway.
|
| |
FEMS Microbiol Lett,
179,
203-208.
|
 |
|
|
|
|
 |
M.Seeger,
M.Zielinski,
K.N.Timmis,
and
B.Hofer
(1999).
Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400.
|
| |
Appl Environ Microbiol,
65,
3614-3621.
|
 |
|
|
|
|
 |
W.S.Somers,
M.L.Stahl,
and
F.X.Sullivan
(1998).
GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site.
|
| |
Structure,
6,
1601-1612.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

