 |
PDBsum entry 1b8s

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.3.6
- cholesterol oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cholesterol + O2 = cholest-5-en-3-one + H2O2
|
 |
 |
 |
 |
 |
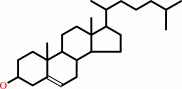 cholesterol
cholesterol
|
+
|
 O2
O2
|
=
|
cholest-5-en-3-one
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.5.3.3.1
- steroid Delta-isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 3-oxo-Delta5-steroid = a 3-oxo-Delta4-steroid
|
 |
 |
 |
 |
 |
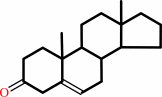 3-oxo-Delta(5)-steroid
3-oxo-Delta(5)-steroid
|
=
|
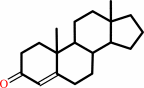 3-oxo-Delta(4)-steroid
3-oxo-Delta(4)-steroid
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
38:4277-4286
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
|
|
Q.K.Yue,
I.J.Kass,
N.S.Sampson,
A.Vrielink.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cholesterol oxidase is a monomeric flavoenzyme which catalyzes the oxidation and
isomerization of cholesterol to cholest-4-en-3-one. The enzyme interacts with
lipid bilayers in order to bind its steroid substrate. The X-ray structure of
the enzyme from Brevibacterium sterolicum revealed two loops, comprising
residues 78-87 and residues 433-436, which act as a lid over the active site and
facilitate binding of the substrate [Vrielink et al. (1991) J. Mol. Biol. 219,
533-554; Li et al. (1993) Biochemistry 32, 11507-11515]. It was postulated that
these loops must open, forming a hydrophobic channel between the membrane and
the active site of the protein and thus sequestering the cholesterol substrate
from the aqueous environment. Here we describe the three-dimensional structure
of the homologous enzyme from Streptomyces refined to 1.5 A resolution.
Structural comparisons to the enzyme from B. sterolicum reveal significant
conformational differences in these loop regions; in particular, a region of the
loop comprising residues 78-87 adopts a small amphipathic helical turn with
hydrophobic residues directed toward the active site cavity and hydrophilic
residues directed toward the external surface of the molecule. It seems
reasonable that this increased rigidity reduces the entropy loss that occurs
upon binding substrate. Consequently, the Streptomyces enzyme is a more
efficient catalyst. In addition, we have determined the structures of three
active site mutants which have significantly reduced activity for either the
oxidation (His447Asn and His447Gln) or the isomerization (Glu361Gln). Our
structural and kinetic data indicate that His447 and Glu361 act as general base
catalysts in association with conserved water H2O541 and Asn485. The His447,
Glu361, H2O541, and Asn485 hydrogen bond network is conserved among other
oxidoreductases. This catalytic tetrad appears to be a structural motif that
occurs in flavoenzymes that catalyze the oxidation of unactivated alcohols.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Uhía,
B.Galán,
V.Morales,
and
J.L.García
(2011).
Initial step in the catabolism of cholesterol by Mycobacterium smegmatis mc2 155.
|
| |
Environ Microbiol,
13,
943-959.
|
 |
|
|
|
|
 |
Y.Xin,
H.Yang,
X.Xia,
L.Zhang,
C.Cheng,
G.Mou,
J.Shi,
Y.Han,
and
W.Wang
(2011).
Affinity purification of a cholesterol oxidase expressed in Escherichia coli.
|
| |
J Chromatogr B Analyt Technol Biomed Life Sci,
879,
853-858.
|
 |
|
|
|
|
 |
A.Y.Lyubimov,
L.Chen,
N.S.Sampson,
and
A.Vrielink
(2009).
A hydrogen-bonding network is important for oxidation and isomerization in the reaction catalyzed by cholesterol oxidase.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1222-1231.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Kreit,
and
N.S.Sampson
(2009).
Cholesterol oxidase: physiological functions.
|
| |
FEBS J,
276,
6844-6856.
|
 |
|
|
|
|
 |
N.Doukyu
(2009).
Characteristics and biotechnological applications of microbial cholesterol oxidases.
|
| |
Appl Microbiol Biotechnol,
83,
825-837.
|
 |
|
|
|
|
 |
J.F.Aparicio,
and
J.F.Martín
(2008).
Microbial cholesterol oxidases: bioconversion enzymes or signal proteins?
|
| |
Mol Biosyst,
4,
804-809.
|
 |
|
|
|
|
 |
J.I.Yeh,
U.Chinte,
and
S.Du
(2008).
Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
|
| |
Proc Natl Acad Sci U S A,
105,
3280-3285.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.K.Arya,
M.Datta,
and
B.D.Malhotra
(2008).
Recent advances in cholesterol biosensor.
|
| |
Biosens Bioelectron,
23,
1083-1100.
|
 |
|
|
|
|
 |
A.Y.Lyubimov,
K.Heard,
H.Tang,
N.S.Sampson,
and
A.Vrielink
(2007).
Distortion of flavin geometry is linked to ligand binding in cholesterol oxidase.
|
| |
Protein Sci,
16,
2647-2656.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Gimpl,
and
K.Gehrig-Burger
(2007).
Cholesterol reporter molecules.
|
| |
Biosci Rep,
27,
335-358.
|
 |
|
|
|
|
 |
M.V.Mendes,
E.Recio,
N.Antón,
S.M.Guerra,
J.Santos-Aberturas,
J.F.Martín,
and
J.F.Aparicio
(2007).
Cholesterol oxidases act as signaling proteins for the biosynthesis of the polyene macrolide pimaricin.
|
| |
Chem Biol,
14,
279-290.
|
 |
|
|
|
|
 |
N.M.Nesbitt,
and
N.S.Sampson
(2007).
Antifungal tradecraft by cholesterol oxidase.
|
| |
Chem Biol,
14,
238-241.
|
 |
|
|
|
|
 |
A.Y.Lyubimov,
P.I.Lario,
I.Moustafa,
and
A.Vrielink
(2006).
Atomic resolution crystallography reveals how changes in pH shape the protein microenvironment.
|
| |
Nat Chem Biol,
2,
259-264.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Ferreira,
F.J.Ruiz-Dueñas,
M.J.Martínez,
W.J.van Berkel,
and
A.T.Martínez
(2006).
Site-directed mutagenesis of selected residues at the active site of aryl-alcohol oxidase, an H2O2-producing ligninolytic enzyme.
|
| |
FEBS J,
273,
4878-4888.
|
 |
|
|
|
|
 |
G.K.Kouassi,
J.Irudayaraj,
and
G.McCarty
(2005).
Examination of Cholesterol oxidase attachment to magnetic nanoparticles.
|
| |
J Nanobiotechnology,
3,
1.
|
 |
|
|
|
|
 |
L.Caldinelli,
S.Iametti,
A.Barbiroli,
F.Bonomi,
D.Fessas,
G.Molla,
M.S.Pilone,
and
L.Pollegioni
(2005).
Dissecting the structural determinants of the stability of cholesterol oxidase containing covalently bound flavin.
|
| |
J Biol Chem,
280,
22572-22581.
|
 |
|
|
|
|
 |
R.D.Hayward,
R.J.Cain,
E.J.McGhie,
N.Phillips,
M.J.Garner,
and
V.Koronakis
(2005).
Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells.
|
| |
Mol Microbiol,
56,
590-603.
|
 |
|
|
|
|
 |
Y.S.Yun,
G.H.Nam,
Y.G.Kim,
B.H.Oh,
and
K.Y.Choi
(2005).
Small exterior hydrophobic cluster contributes to conformational stability and steroid binding in ketosteroid isomerase from Pseudomonas putida biotype B.
|
| |
FEBS J,
272,
1999-2011.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Vrielink,
and
N.Sampson
(2003).
Sub-Angstrom resolution enzyme X-ray structures: is seeing believing?
|
| |
Curr Opin Struct Biol,
13,
709-715.
|
 |
|
|
|
|
 |
N.Friedland,
H.L.Liou,
P.Lobel,
and
A.M.Stock
(2003).
Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease.
|
| |
Proc Natl Acad Sci U S A,
100,
2512-2517.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.A.Bottoms,
P.E.Smith,
and
J.J.Tanner
(2002).
A structurally conserved water molecule in Rossmann dinucleotide-binding domains.
|
| |
Protein Sci,
11,
2125-2137.
|
 |
|
|
|
|
 |
I.Dreveny,
C.Kratky,
and
K.Gruber
(2002).
The active site of hydroxynitrile lyase from Prunus amygdalus: modeling studies provide new insights into the mechanism of cyanogenesis.
|
| |
Protein Sci,
11,
292-300.
|
 |
|
|
|
|
 |
R.Aunpad,
S.P.Muench,
P.J.Baker,
S.Sedelnikova,
W.Panbangred,
N.Doukyu,
R.Aono,
and
D.W.Rice
(2002).
Crystallization and preliminary X-ray crystallographic studies on the class II cholesterol oxidase from Burkholderia cepacia containing bound flavin.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
2182-2183.
|
 |
|
|
|
|
 |
N.S.Sampson
(2001).
Dissection of a flavoenzyme active site: the reaction catalyzed by cholesterol oxidase.
|
| |
Antioxid Redox Signal,
3,
839-846.
|
 |
|
|
|
|
 |
O.Dym,
and
D.Eisenberg
(2001).
Sequence-structure analysis of FAD-containing proteins.
|
| |
Protein Sci,
10,
1712-1728.
|
 |
|
|
|
|
 |
X.Chen,
D.E.Wolfgang,
and
N.S.Sampson
(2000).
Use of the parallax-quench method to determine the position of the active-site loop of cholesterol oxidase in lipid bilayers.
|
| |
Biochemistry,
39,
13383-13389.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

