 |
PDBsum entry 1b5o

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.1
- aspartate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate
|
 |
 |
 |
 |
 |
 L-aspartate
L-aspartate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 oxaloacetate
oxaloacetate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.78
- aspartate--prephenate aminotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-arogenate + oxaloacetate = prephenate + L-aspartate
|
 |
 |
 |
 |
 |
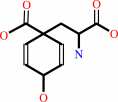 L-arogenate
L-arogenate
|
+
|
 oxaloacetate
oxaloacetate
|
=
|
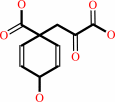 prephenate
prephenate
|
+
|
 L-aspartate
L-aspartate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biochem (tokyo)
130:89-98
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Substrate recognition mechanism of thermophilic dual-substrate enzyme.
|
|
H.Ura,
T.Nakai,
S.I.Kawaguchi,
I.Miyahara,
K.Hirotsu,
S.Kuramitsu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aspartate aminotransferase from an extremely thermophilic bacterium, Thermus
thermophilus HB8 (ttAspAT), has been believed to be specific for an acidic
substrate. However, stepwise introduction of mutations in the active-site
residues finally changed its substrate specificity to that of a dual-substrate
enzyme. The final mutant, [S15D, T17V, K109S, S292R] ttAspAT, is active toward
both acidic and hydrophobic substrates. During the course of stepwise mutation,
the activities toward acidic and hydrophobic substrates changed independently.
The introduction of a mobile Arg292* residue into ttAspAT was the key step in
the change to a "dual-substrate" enzyme. The substrate recognition mechanism of
this thermostable "dual-substrate" enzyme was confirmed by X-ray
crystallography. This work together with previous studies on various enzymes
suggest that this unique "dual-substrate recognition" mechanism is a feature of
not only aminotransferases but also other enzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.P.Dubnovitsky,
E.G.Kapetaniou,
and
A.C.Papageorgiou
(2005).
Enzyme adaptation to alkaline pH: atomic resolution (1.08 A) structure of phosphoserine aminotransferase from Bacillus alcalophilus.
|
| |
Protein Sci,
14,
97.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.P.Dubnovitsky,
R.B.Ravelli,
A.N.Popov,
and
A.C.Papageorgiou
(2005).
Strain relief at the active site of phosphoserine aminotransferase induced by radiation damage.
|
| |
Protein Sci,
14,
1498-1507.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Hirotsu,
M.Goto,
A.Okamoto,
and
I.Miyahara
(2005).
Dual substrate recognition of aminotransferases.
|
| |
Chem Rec,
5,
160-172.
|
 |
|
|
|
|
 |
S.Yoshiba,
N.Nakagawa,
R.Masui,
T.Shibata,
Y.Inoue,
S.Yokoyama,
and
S.Kuramitsu
(2003).
Overproduction, crystallization and preliminary diffraction data of ADP-ribose pyrophosphatase from Thermus thermophilus HB8.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1840-1842.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

