 |
PDBsum entry 1amu

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Peptide synthetase
|
PDB id
|
|

|

|
1amu
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.1.11
- phenylalanine racemase (ATP-hydrolyzing).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-phenylalanine + ATP + H2O = D-phenylalanine + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
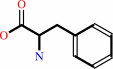 L-phenylalanine
L-phenylalanine
|
+
|
 ATP
ATP
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
D-phenylalanine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Embo J
16:4174-4183
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S.
|
|
E.Conti,
T.Stachelhaus,
M.A.Marahiel,
P.Brick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The non-ribosomal synthesis of the cyclic peptide antibiotic gramicidin S is
accomplished by two large multifunctional enzymes, the peptide synthetases 1 and
2. The enzyme complex contains five conserved subunits of approximately 60 kDa
which carry out ATP-dependent activation of specific amino acids and share
extensive regions of sequence similarity with adenylating enzymes such as
firefly luciferases and acyl-CoA ligases. We have determined the crystal
structure of the N-terminal adenylation subunit in a complex with AMP and
L-phenylalanine to 1.9 A resolution. The 556 amino acid residue fragment is
folded into two domains with the active site situated at their interface. Each
domain of the enzyme has a similar topology to the corresponding domain of
unliganded firefly luciferase, but a remarkable relative domain rotation of 94
degrees occurs. This conformation places the absolutely conserved Lys517 in a
position to form electrostatic interactions with both ligands. The AMP is bound
with the phosphate moiety interacting with Lys517 and the hydroxyl groups of the
ribose forming hydrogen bonds with Asp413. The phenylalanine substrate binds in
a hydrophobic pocket with the carboxylate group interacting with Lys517 and the
alpha-amino group with Asp235. The structure reveals the role of the invariant
residues within the superfamily of adenylate-forming enzymes and indicates a
conserved mechanism of nucleotide binding and substrate activation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2 Stereo diagram of the large N-terminal domain of PheA
showing the bound ligands coloured as in Figure 1. The  -sheet
A is on the left-hand side, -sheet
A is on the left-hand side,  -sheet
B is on the right-hand side, and the -sheet
B is on the right-hand side, and the  -barrel
is at the top of the figure. The N-terminus of the protein is at
the top of the figure. The disordered loop (residues 192 -196)
near the active site is coloured violet. -barrel
is at the top of the figure. The N-terminus of the protein is at
the top of the figure. The disordered loop (residues 192 -196)
near the active site is coloured violet.
|
 |
Figure 6.
Figure 6 Schematic representation of the hydrogen bonding
between PheA and the phenylalanine and AMP ligands.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
Embo J
(1997,
16,
4174-4183)
copyright 1997.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.J.Hughes,
and
A.Keatinge-Clay
(2011).
Enzymatic extender unit generation for in vitro polyketide synthase reactions: structural and functional showcasing of Streptomyces coelicolor MatB.
|
| |
Chem Biol,
18,
165-176.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Höfer,
M.Crüsemann,
M.Radzom,
B.Geers,
D.Flachshaar,
X.Cai,
A.Zeeck,
and
J.Piel
(2011).
Insights into the biosynthesis of hormaomycin, an exceptionally complex bacterial signaling metabolite.
|
| |
Chem Biol,
18,
381-391.
|
 |
|
|
|
|
 |
M.J.Koetsier,
P.A.Jekel,
H.J.Wijma,
R.A.Bovenberg,
and
D.B.Janssen
(2011).
Aminoacyl-coenzyme A synthesis catalyzed by a CoA ligase from Penicillium chrysogenum.
|
| |
FEBS Lett,
585,
893-898.
|
 |
|
|
|
|
 |
R.Süssmuth,
J.Müller,
H.von Döhren,
and
I.Molnár
(2011).
Fungal cyclooligomer depsipeptides: from classical biochemistry to combinatorial biosynthesis.
|
| |
Nat Prod Rep,
28,
99.
|
 |
|
|
|
|
 |
S.Hosseinkhani
(2011).
Molecular enigma of multicolor bioluminescence of firefly luciferase.
|
| |
Cell Mol Life Sci,
68,
1167-1182.
|
 |
|
|
|
|
 |
B.D.Ames,
and
C.T.Walsh
(2010).
Anthranilate-activating modules from fungal nonribosomal peptide assembly lines.
|
| |
Biochemistry,
49,
3351-3365.
|
 |
|
|
|
|
 |
C.Wu,
R.Cichewicz,
Y.Li,
J.Liu,
B.Roe,
J.Ferretti,
J.Merritt,
and
F.Qi
(2010).
Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance.
|
| |
Appl Environ Microbiol,
76,
5815-5826.
|
 |
|
|
|
|
 |
H.A.Crosby,
E.K.Heiniger,
C.S.Harwood,
and
J.C.Escalante-Semerena
(2010).
Reversible N epsilon-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris.
|
| |
Mol Microbiol,
76,
874-888.
|
 |
|
|
|
|
 |
K.Fujiwara,
N.Maita,
H.Hosaka,
K.Okamura-Ikeda,
A.Nakagawa,
and
H.Taniguchi
(2010).
Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A.
|
| |
J Biol Chem,
285,
9971-9980.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Anand,
M.V.Prasad,
G.Yadav,
N.Kumar,
J.Shehara,
M.Z.Ansari,
and
D.Mohanty
(2010).
SBSPKS: structure based sequence analysis of polyketide synthases.
|
| |
Nucleic Acids Res,
38,
W487-W496.
|
 |
|
|
|
|
 |
T.V.Lee,
L.J.Johnson,
R.D.Johnson,
A.Koulman,
G.A.Lane,
J.S.Lott,
and
V.L.Arcus
(2010).
Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis.
|
| |
J Biol Chem,
285,
2415-2427.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Koglin,
and
C.T.Walsh
(2009).
Structural insights into nonribosomal peptide enzymatic assembly lines.
|
| |
Nat Prod Rep,
26,
987.
|
 |
|
|
|
|
 |
A.M.Gulick
(2009).
Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase.
|
| |
ACS Chem Biol,
4,
811-827.
|
 |
|
|
|
|
 |
B.M.Kevany,
D.A.Rasko,
and
M.G.Thomas
(2009).
Characterization of the complete zwittermicin A biosynthesis gene cluster from Bacillus cereus.
|
| |
Appl Environ Microbiol,
75,
1144-1155.
|
 |
|
|
|
|
 |
B.R.Villiers,
and
F.Hollfelder
(2009).
Mapping the limits of substrate specificity of the adenylation domain of TycA.
|
| |
Chembiochem,
10,
671-682.
|
 |
|
|
|
|
 |
C.Y.Chen,
I.Georgiev,
A.C.Anderson,
and
B.R.Donald
(2009).
Computational structure-based redesign of enzyme activity.
|
| |
Proc Natl Acad Sci U S A,
106,
3764-3769.
|
 |
|
|
|
|
 |
H.Jenke-Kodama,
and
E.Dittmann
(2009).
Bioinformatic perspectives on NRPS/PKS megasynthases: Advances and challenges.
|
| |
Nat Prod Rep,
26,
874-883.
|
 |
|
|
|
|
 |
J.Zaitseva,
K.M.Meneely,
and
A.L.Lamb
(2009).
Structure of Escherichia coli malate dehydrogenase at 1.45 A resolution.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
866-869.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.B.Shah,
C.Ingram-Smith,
L.L.Cooper,
J.Qu,
Y.Meng,
K.S.Smith,
and
A.M.Gulick
(2009).
The 2.1 A crystal structure of an acyl-CoA synthetase from Methanosarcina acetivorans reveals an alternate acyl-binding pocket for small branched acyl substrates.
|
| |
Proteins,
77,
685-698.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Arora,
A.Goyal,
V.T.Natarajan,
E.Rajakumara,
P.Verma,
R.Gupta,
M.Yousuf,
O.A.Trivedi,
D.Mohanty,
A.Tyagi,
R.Sankaranarayanan,
and
R.S.Gokhale
(2009).
Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis.
|
| |
Nat Chem Biol,
5,
166-173.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Zhu,
Y.Zheng,
Y.You,
X.Yan,
and
J.Shao
(2009).
Sequencing and modular analysis of the hybrid non-ribosomal peptide synthase - polyketide synthase gene cluster from the marine sponge Hymeniacidon perleve-associated bacterium Pseudoalteromonas sp. strain NJ631.
|
| |
Can J Microbiol,
55,
219-227.
|
 |
|
|
|
|
 |
R.Wu,
A.S.Reger,
X.Lu,
A.M.Gulick,
and
D.Dunaway-Mariano
(2009).
The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase.
|
| |
Biochemistry,
48,
4115-4125.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Schmelz,
N.Kadi,
S.A.McMahon,
L.Song,
D.Oves-Costales,
M.Oke,
H.Liu,
K.A.Johnson,
L.G.Carter,
C.H.Botting,
M.F.White,
G.L.Challis,
and
J.H.Naismith
(2009).
AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis.
|
| |
Nat Chem Biol,
5,
174-182.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.J.Ke,
B.Y.Chang,
T.P.Lin,
and
S.T.Liu
(2009).
Activation of the promoter of the fengycin synthetase operon by the UP element.
|
| |
J Bacteriol,
191,
4615-4623.
|
 |
|
|
|
|
 |
A.Endler,
S.Martens,
F.Wellmann,
and
U.Matern
(2008).
Unusually divergent 4-coumarate:CoA-ligases from Ruta graveolens L.
|
| |
Plant Mol Biol,
67,
335-346.
|
 |
|
|
|
|
 |
A.Koglin,
F.Löhr,
F.Bernhard,
V.V.Rogov,
D.P.Frueh,
E.R.Strieter,
M.R.Mofid,
P.Güntert,
G.Wagner,
C.T.Walsh,
M.A.Marahiel,
and
V.Dötsch
(2008).
Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
|
| |
Nature,
454,
907-911.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.S.Reger,
R.Wu,
D.Dunaway-Mariano,
and
A.M.Gulick
(2008).
Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
|
| |
Biochemistry,
47,
8016-8025.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Tanovic,
S.A.Samel,
L.O.Essen,
and
M.A.Marahiel
(2008).
Crystal structure of the termination module of a nonribosomal peptide synthetase.
|
| |
Science,
321,
659-663.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Tooming-Klunderud,
D.P.Fewer,
T.Rohrlack,
J.Jokela,
L.Rouhiainen,
K.Sivonen,
T.Kristensen,
and
K.S.Jakobsen
(2008).
Evidence for positive selection acting on microcystin synthetase adenylation domains in three cyanobacterial genera.
|
| |
BMC Evol Biol,
8,
256.
|
 |
|
|
|
|
 |
C.T.Calderone,
S.B.Bumpus,
N.L.Kelleher,
C.T.Walsh,
and
N.A.Magarvey
(2008).
A ketoreductase domain in the PksJ protein of the bacillaene assembly line carries out both alpha- and beta-ketone reduction during chain growth.
|
| |
Proc Natl Acad Sci U S A,
105,
12809-12814.
|
 |
|
|
|
|
 |
G.Christiansen,
W.Y.Yoshida,
J.F.Blom,
C.Portmann,
K.Gademann,
T.Hemscheidt,
and
R.Kurmayer
(2008).
Isolation and Structure Determination of Two Microcystins and Sequence Comparison of the McyABC Adenylation Domains in Planktothrix Species.
|
| |
J Nat Prod,
71,
1881-1886.
|
 |
|
|
|
|
 |
G.D.Amoutzias,
Y.Van de Peer,
and
D.Mossialos
(2008).
Evolution and taxonomic distribution of nonribosomal peptide and polyketide synthases.
|
| |
Future Microbiol,
3,
361-370.
|
 |
|
|
|
|
 |
G.Mercado,
M.Tello,
M.Marín,
O.Monasterio,
and
R.Lagos
(2008).
The production in vivo of microcin E492 with antibacterial activity depends on salmochelin and EntF.
|
| |
J Bacteriol,
190,
5464-5471.
|
 |
|
|
|
|
 |
H.Fraga
(2008).
Firefly luminescence: a historical perspective and recent developments.
|
| |
Photochem Photobiol Sci,
7,
146-158.
|
 |
|
|
|
|
 |
H.Haas,
M.Eisendle,
and
B.G.Turgeon
(2008).
Siderophores in fungal physiology and virulence.
|
| |
Annu Rev Phytopathol,
46,
149-187.
|
 |
|
|
|
|
 |
H.Yonus,
P.Neumann,
S.Zimmermann,
J.J.May,
M.A.Marahiel,
and
M.T.Stubbs
(2008).
Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains.
|
| |
J Biol Chem,
283,
32484-32491.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Georgiev,
D.Keedy,
J.S.Richardson,
D.C.Richardson,
and
B.R.Donald
(2008).
Algorithm for backrub motions in protein design.
|
| |
Bioinformatics,
24,
i196-i204.
|
 |
|
|
|
|
 |
I.Georgiev,
R.H.Lilien,
and
B.R.Donald
(2008).
The minimized dead-end elimination criterion and its application to protein redesign in a hybrid scoring and search algorithm for computing partition functions over molecular ensembles.
|
| |
J Comput Chem,
29,
1527-1542.
|
 |
|
|
|
|
 |
J.D.Awaya,
and
J.L.Dubois
(2008).
Identification, isolation, and analysis of a gene cluster involved in iron acquisition by Pseudomonas mendocina ymp.
|
| |
Biometals,
21,
353-366.
|
 |
|
|
|
|
 |
J.S.Cisar,
and
D.S.Tan
(2008).
Small molecule inhibition of microbial natural product biosynthesis-an emerging antibiotic strategy.
|
| |
Chem Soc Rev,
37,
1320-1329.
|
 |
|
|
|
|
 |
K.E.Bushley,
D.R.Ripoll,
and
B.G.Turgeon
(2008).
Module evolution and substrate specificity of fungal nonribosomal peptide synthetases involved in siderophore biosynthesis.
|
| |
BMC Evol Biol,
8,
328.
|
 |
|
|
|
|
 |
K.J.Weissman,
and
R.Müller
(2008).
Protein-protein interactions in multienzyme megasynthetases.
|
| |
Chembiochem,
9,
826-848.
|
 |
|
|
|
|
 |
L.M.Halo,
J.W.Marshall,
A.A.Yakasai,
Z.Song,
C.P.Butts,
M.P.Crump,
M.Heneghan,
A.M.Bailey,
T.J.Simpson,
C.M.Lazarus,
and
R.J.Cox
(2008).
Authentic heterologous expression of the tenellin iterative polyketide synthase nonribosomal peptide synthetase requires coexpression with an enoyl reductase.
|
| |
Chembiochem,
9,
585-594.
|
 |
|
|
|
|
 |
T.Abe,
Y.Hashimoto,
H.Hosaka,
K.Tomita-Yokotani,
and
M.Kobayashi
(2008).
Discovery of amide (peptide) bond synthetic activity in Acyl-CoA synthetase.
|
| |
J Biol Chem,
283,
11312-11321.
|
 |
|
|
|
|
 |
X.Lu,
H.Zhang,
P.J.Tonge,
and
D.S.Tan
(2008).
Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis.
|
| |
Bioorg Med Chem Lett,
18,
5963-5966.
|
 |
|
|
|
|
 |
A.D.Berti,
N.J.Greve,
Q.H.Christensen,
and
M.G.Thomas
(2007).
Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000.
|
| |
J Bacteriol,
189,
6312-6323.
|
 |
|
|
|
|
 |
A.Renier,
E.Vivien,
S.Cociancich,
P.Letourmy,
X.Perrier,
P.C.Rott,
and
M.Royer
(2007).
Substrate specificity-conferring regions of the nonribosomal peptide synthetase adenylation domains involved in albicidin pathotoxin biosynthesis are highly conserved within the species Xanthomonas albilineans.
|
| |
Appl Environ Microbiol,
73,
5523-5530.
|
 |
|
|
|
|
 |
A.S.Reger,
J.M.Carney,
and
A.M.Gulick
(2007).
Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase.
|
| |
Biochemistry,
46,
6536-6546.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Szarecka,
Y.Xu,
and
P.Tang
(2007).
Dynamics of firefly luciferase inhibition by general anesthetics: Gaussian and anisotropic network analyses.
|
| |
Biophys J,
93,
1895-1905.
|
 |
|
|
|
|
 |
C.Ingram-Smith,
and
K.S.Smith
(2007).
AMP-forming acetyl-CoA synthetases in Archaea show unexpected diversity in substrate utilization.
|
| |
Archaea,
2,
95.
|
 |
|
|
|
|
 |
C.J.Balibar,
A.R.Howard-Jones,
and
C.T.Walsh
(2007).
Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation.
|
| |
Nat Chem Biol,
3,
584-592.
|
 |
|
|
|
|
 |
C.Y.Wu,
C.L.Chen,
Y.H.Lee,
Y.C.Cheng,
Y.C.Wu,
H.Y.Shu,
F.Götz,
and
S.T.Liu
(2007).
Nonribosomal synthesis of fengycin on an enzyme complex formed by fengycin synthetases.
|
| |
J Biol Chem,
282,
5608-5616.
|
 |
|
|
|
|
 |
H.Takahashi,
T.Kumagai,
K.Kitani,
M.Mori,
Y.Matoba,
and
M.Sugiyama
(2007).
Cloning and characterization of a Streptomyces single module type non-ribosomal peptide synthetase catalyzing a blue pigment synthesis.
|
| |
J Biol Chem,
282,
9073-9081.
|
 |
|
|
|
|
 |
I.Georgiev,
and
B.R.Donald
(2007).
Dead-end elimination with backbone flexibility.
|
| |
Bioinformatics,
23,
i185-i194.
|
 |
|
|
|
|
 |
J.S.Cisar,
J.A.Ferreras,
R.K.Soni,
L.E.Quadri,
and
D.S.Tan
(2007).
Exploiting ligand conformation in selective inhibition of non-ribosomal peptide synthetase amino acid adenylation with designed macrocyclic small molecules.
|
| |
J Am Chem Soc,
129,
7752-7753.
|
 |
|
|
|
|
 |
K.L.Eley,
L.M.Halo,
Z.Song,
H.Powles,
R.J.Cox,
A.M.Bailey,
C.M.Lazarus,
and
T.J.Simpson
(2007).
Biosynthesis of the 2-pyridone tenellin in the insect pathogenic fungus Beauveria bassiana.
|
| |
Chembiochem,
8,
289-297.
|
 |
|
|
|
|
 |
L.G.Otten,
M.L.Schaffer,
B.R.Villiers,
T.Stachelhaus,
and
F.Hollfelder
(2007).
An optimized ATP/PP(i)-exchange assay in 96-well format for screening of adenylation domains for applications in combinatorial biosynthesis.
|
| |
Biotechnol J,
2,
232-240.
|
 |
|
|
|
|
 |
P.Schneider,
M.Weber,
K.Rosenberger,
and
D.Hoffmeister
(2007).
A one-pot chemoenzymatic synthesis for the universal precursor of antidiabetes and antiviral bis-indolylquinones.
|
| |
Chem Biol,
14,
635-644.
|
 |
|
|
|
|
 |
R.S.Gokhale,
P.Saxena,
T.Chopra,
and
D.Mohanty
(2007).
Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids.
|
| |
Nat Prod Rep,
24,
267-277.
|
 |
|
|
|
|
 |
S.P.Lim,
N.Roongsawang,
K.Washio,
and
M.Morikawa
(2007).
Functional analysis of a pyoverdine synthetase from Pseudomonas sp. MIS38.
|
| |
Biosci Biotechnol Biochem,
71,
2002-2009.
|
 |
|
|
|
|
 |
A.V.Demirev,
C.H.Lee,
B.P.Jaishy,
D.H.Nam,
and
D.D.Ryu
(2006).
Substrate specificity of nonribosomal peptide synthetase modules responsible for the biosynthesis of the oligopeptide moiety of cephabacin in Lysobacter lactamgenus.
|
| |
FEMS Microbiol Lett,
255,
121-128.
|
 |
|
|
|
|
 |
D.B.Stein,
U.Linne,
M.Hahn,
and
M.A.Marahiel
(2006).
Impact of epimerization domains on the intermodular transfer of enzyme-bound intermediates in nonribosomal peptide synthesis.
|
| |
Chembiochem,
7,
1807-1814.
|
 |
|
|
|
|
 |
E.Arias-Barrau,
E.R.Olivera,
A.Sandoval,
G.Naharro,
and
J.M.Luengo
(2006).
Acetyl-CoA synthetase from Pseudomonas putida U is the only acyl-CoA activating enzyme induced by acetate in this bacterium.
|
| |
FEMS Microbiol Lett,
260,
36-46.
|
 |
|
|
|
|
 |
E.J.Drake,
D.A.Nicolai,
and
A.M.Gulick
(2006).
Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain.
|
| |
Chem Biol,
13,
409-419.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Niu,
G.Liu,
Y.Tian,
and
H.Tan
(2006).
SanJ, an ATP-dependent picolinate-CoA ligase, catalyzes the conversion of picolinate to picolinate-CoA during nikkomycin biosynthesis in Streptomyces ansochromogenes.
|
| |
Metab Eng,
8,
183-195.
|
 |
|
|
|
|
 |
J.Grünewald,
and
M.A.Marahiel
(2006).
Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides.
|
| |
Microbiol Mol Biol Rev,
70,
121-146.
|
 |
|
|
|
|
 |
L.M.Hicks,
C.J.Balibar,
C.T.Walsh,
N.L.Kelleher,
and
N.J.Hillson
(2006).
Probing intra- versus interchain kinetic preferences of L-Thr acylation on dimeric VibF with mass spectrometry.
|
| |
Biophys J,
91,
2609-2619.
|
 |
|
|
|
|
 |
M.Ehling-Schulz,
M.Fricker,
H.Grallert,
P.Rieck,
M.Wagner,
and
S.Scherer
(2006).
Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1.
|
| |
BMC Microbiol,
6,
20.
|
 |
|
|
|
|
 |
R.Kellmann,
T.Mills,
and
B.A.Neilan
(2006).
Functional modeling and phylogenetic distribution of putative cylindrospermopsin biosynthesis enzymes.
|
| |
J Mol Evol,
62,
267-280.
|
 |
|
|
|
|
 |
R.Kurmayer,
and
M.Gumpenberger
(2006).
Diversity of microcystin genotypes among populations of the filamentous cyanobacteria Planktothrix rubescens and Planktothrix agardhii.
|
| |
Mol Ecol,
15,
3849-3861.
|
 |
|
|
|
|
 |
R.V.Somu,
D.J.Wilson,
E.M.Bennett,
H.I.Boshoff,
L.Celia,
B.J.Beck,
C.E.Barry,
and
C.C.Aldrich
(2006).
Antitubercular nucleosides that inhibit siderophore biosynthesis: SAR of the glycosyl domain.
|
| |
J Med Chem,
49,
7623-7635.
|
 |
|
|
|
|
 |
S.C.Wenzel,
P.Meiser,
T.M.Binz,
T.Mahmud,
and
R.Müller
(2006).
Nonribosomal peptide biosynthesis: point mutations and module skipping lead to chemical diversity.
|
| |
Angew Chem Int Ed Engl,
45,
2296-2301.
|
 |
|
|
|
|
 |
S.G.Van Lanen,
S.Lin,
P.C.Dorrestein,
N.L.Kelleher,
and
B.Shen
(2006).
Substrate specificity of the adenylation enzyme SgcC1 involved in the biosynthesis of the enediyne antitumor antibiotic C-1027.
|
| |
J Biol Chem,
281,
29633-29640.
|
 |
|
|
|
|
 |
T.Nakatsu,
S.Ichiyama,
J.Hiratake,
A.Saldanha,
N.Kobashi,
K.Sakata,
and
H.Kato
(2006).
Structural basis for the spectral difference in luciferase bioluminescence.
|
| |
Nature,
440,
372-376.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Q.Cheng
(2006).
Deciphering the biosynthetic codes for the potent anti-SARS-CoV cyclodepsipeptide valinomycin in Streptomyces tsusimaensis ATCC 15141.
|
| |
Chembiochem,
7,
471-477.
|
 |
|
|
|
|
 |
C.J.Balibar,
F.H.Vaillancourt,
and
C.T.Walsh
(2005).
Generation of D amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains.
|
| |
Chem Biol,
12,
1189-1200.
|
 |
|
|
|
|
 |
C.Rausch,
T.Weber,
O.Kohlbacher,
W.Wohlleben,
and
D.H.Huson
(2005).
Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs).
|
| |
Nucleic Acids Res,
33,
5799-5808.
|
 |
|
|
|
|
 |
D.B.Stein,
U.Linne,
and
M.A.Marahiel
(2005).
Utility of epimerization domains for the redesign of nonribosomal peptide synthetases.
|
| |
FEBS J,
272,
4506-4520.
|
 |
|
|
|
|
 |
D.G.Panaccione
(2005).
Origins and significance of ergot alkaloid diversity in fungi.
|
| |
FEMS Microbiol Lett,
251,
9.
|
 |
|
|
|
|
 |
D.T.Huang,
A.Paydar,
M.Zhuang,
M.B.Waddell,
J.M.Holton,
and
B.A.Schulman
(2005).
Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1.
|
| |
Mol Cell,
17,
341-350.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.J.May,
R.Finking,
F.Wiegeshoff,
T.T.Weber,
N.Bandur,
U.Koert,
and
M.A.Marahiel
(2005).
Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium's susceptibility to antibiotics that target the cell wall.
|
| |
FEBS J,
272,
2993-3003.
|
 |
|
|
|
|
 |
J.Raap,
K.Erkelens,
A.Ogrel,
D.A.Skladnev,
and
H.Brückner
(2005).
Fungal biosynthesis of non-ribosomal peptide antibiotics and alpha, alpha-dialkylated amino acid constituents.
|
| |
J Pept Sci,
11,
331-338.
|
 |
|
|
|
|
 |
N.N.Ugarova,
L.G.Maloshenok,
I.V.Uporov,
and
M.I.Koksharov
(2005).
Bioluminescence spectra of native and mutant firefly luciferases as a function of pH.
|
| |
Biochemistry (Mosc),
70,
1262-1267.
|
 |
|
|
|
|
 |
R.H.Lilien,
B.W.Stevens,
A.C.Anderson,
and
B.R.Donald
(2005).
A novel ensemble-based scoring and search algorithm for protein redesign and its application to modify the substrate specificity of the gramicidin synthetase a phenylalanine adenylation enzyme.
|
| |
J Comput Biol,
12,
740-761.
|
 |
|
|
|
|
 |
S.C.Wenzel,
B.Kunze,
G.Höfle,
B.Silakowski,
M.Scharfe,
H.Blöcker,
and
R.Müller
(2005).
Structure and biosynthesis of myxochromides S1-3 in Stigmatella aurantiaca: evidence for an iterative bacterial type I polyketide synthase and for module skipping in nonribosomal peptide biosynthesis.
|
| |
Chembiochem,
6,
375-385.
|
 |
|
|
|
|
 |
S.K.Samanta,
and
C.S.Harwood
(2005).
Use of the Rhodopseudomonas palustris genome sequence to identify a single amino acid that contributes to the activity of a coenzyme A ligase with chlorinated substrates.
|
| |
Mol Microbiol,
55,
1151-1159.
|
 |
|
|
|
|
 |
X.Wu,
J.Ballard,
and
Y.W.Jiang
(2005).
Structure and biosynthesis of the BT peptide antibiotic from Brevibacillus texasporus.
|
| |
Appl Environ Microbiol,
71,
8519-8530.
|
 |
|
|
|
|
 |
Y.Hashimoto,
H.Hosaka,
K.Oinuma,
M.Goda,
H.Higashibata,
and
M.Kobayashi
(2005).
Nitrile pathway involving acyl-CoA synthetase: overall metabolic gene organization and purification and characterization of the enzyme.
|
| |
J Biol Chem,
280,
8660-8667.
|
 |
|
|
|
|
 |
Y.Li,
N.M.Llewellyn,
R.Giri,
F.Huang,
and
J.B.Spencer
(2005).
Biosynthesis of the unique amino acid side chain of butirosin: possible protective-group chemistry in an acyl carrier protein-mediated pathway.
|
| |
Chem Biol,
12,
665-675.
|
 |
|
|
|
|
 |
Y.Oba,
M.Sato,
M.Ojika,
and
S.Inouye
(2005).
Enzymatic and genetic characterization of firefly luciferase and Drosophila CG6178 as a fatty acyl-CoA synthetase.
|
| |
Biosci Biotechnol Biochem,
69,
819-828.
|
 |
|
|
|
|
 |
A.Sandmann,
F.Sasse,
and
R.Müller
(2004).
Identification and analysis of the core biosynthetic machinery of tubulysin, a potent cytotoxin with potential anticancer activity.
|
| |
Chem Biol,
11,
1071-1079.
|
 |
|
|
|
|
 |
G.L.Tang,
Y.Q.Cheng,
and
B.Shen
(2004).
Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase.
|
| |
Chem Biol,
11,
33-45.
|
 |
|
|
|
|
 |
M.Di Lorenzo,
S.Poppelaars,
M.Stork,
M.Nagasawa,
M.E.Tolmasky,
and
J.H.Crosa
(2004).
A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibrio anguillarum.
|
| |
J Bacteriol,
186,
7327-7336.
|
 |
|
|
|
|
 |
M.Z.Ansari,
G.Yadav,
R.S.Gokhale,
and
D.Mohanty
(2004).
NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases.
|
| |
Nucleic Acids Res,
32,
W405-W413.
|
 |
|
|
|
|
 |
N.Kessler,
H.Schuhmann,
S.Morneweg,
U.Linne,
and
M.A.Marahiel
(2004).
The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains.
|
| |
J Biol Chem,
279,
7413-7419.
|
 |
|
|
|
|
 |
R.Finking,
and
M.A.Marahiel
(2004).
Biosynthesis of nonribosomal peptides1.
|
| |
Annu Rev Microbiol,
58,
453-488.
|
 |
|
|
|
|
 |
R.M.Morgan-Kiss,
and
J.E.Cronan
(2004).
The Escherichia coli fadK (ydiD) gene encodes an anerobically regulated short chain acyl-CoA synthetase.
|
| |
J Biol Chem,
279,
37324-37333.
|
 |
|
|
|
|
 |
T.Duerfahrt,
K.Eppelmann,
R.Müller,
and
M.A.Marahiel
(2004).
Rational design of a bimodular model system for the investigation of heterocyclization in nonribosomal peptide biosynthesis.
|
| |
Chem Biol,
11,
261-271.
|
 |
|
|
|
|
 |
Y.Hisanaga,
H.Ago,
N.Nakagawa,
K.Hamada,
K.Ida,
M.Yamamoto,
T.Hori,
Y.Arii,
M.Sugahara,
S.Kuramitsu,
S.Yokoyama,
and
M.Miyano
(2004).
Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer.
|
| |
J Biol Chem,
279,
31717-31726.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Paradkar,
A.Trefzer,
R.Chakraburtty,
and
D.Stassi
(2003).
Streptomyces genetics: a genomic perspective.
|
| |
Crit Rev Biotechnol,
23,
1.
|
 |
|
|
|
|
 |
A.Richardt,
T.Kemme,
S.Wagner,
D.Schwarzer,
M.A.Marahiel,
and
B.T.Hovemann
(2003).
Ebony, a novel nonribosomal peptide synthetase for beta-alanine conjugation with biogenic amines in Drosophila.
|
| |
J Biol Chem,
278,
41160-41166.
|
 |
|
|
|
|
 |
B.K.Scholz-Schroeder,
J.D.Soule,
and
D.C.Gross
(2003).
The sypA, sypS, and sypC synthetase genes encode twenty-two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D.
|
| |
Mol Plant Microbe Interact,
16,
271-280.
|
 |
|
|
|
|
 |
B.Mikalsen,
G.Boison,
O.M.Skulberg,
J.Fastner,
W.Davies,
T.M.Gabrielsen,
K.Rudi,
and
K.S.Jakobsen
(2003).
Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains.
|
| |
J Bacteriol,
185,
2774-2785.
|
 |
|
|
|
|
 |
C.Lindermayr,
J.Fliegmann,
and
J.Ebel
(2003).
Deletion of a single amino acid residue from different 4-coumarate:CoA ligases from soybean results in the generation of new substrate specificities.
|
| |
J Biol Chem,
278,
2781-2786.
|
 |
|
|
|
|
 |
D.F.Ackerley,
T.T.Caradoc-Davies,
and
I.L.Lamont
(2003).
Substrate specificity of the nonribosomal peptide synthetase PvdD from Pseudomonas aeruginosa.
|
| |
J Bacteriol,
185,
2848-2855.
|
 |
|
|
|
|
 |
K.Schneider,
K.Hövel,
K.Witzel,
B.Hamberger,
D.Schomburg,
E.Kombrink,
and
H.P.Stuible
(2003).
The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase.
|
| |
Proc Natl Acad Sci U S A,
100,
8601-8606.
|
 |
|
|
|
|
 |
L.J.Ming
(2003).
Structure and function of "metalloantibiotics".
|
| |
Med Res Rev,
23,
697-762.
|
 |
|
|
|
|
 |
M.J.Hijarrubia,
J.F.Aparicio,
and
J.F.Martín
(2003).
Domain structure characterization of the multifunctional alpha-aminoadipate reductase from Penicillium chrysogenum by limited proteolysis. Activation of alpha-aminoadipate does not require the peptidyl carrier protein box or the reduction domain.
|
| |
J Biol Chem,
278,
8250-8256.
|
 |
|
|
|
|
 |
M.Mukherji,
C.J.Schofield,
A.S.Wierzbicki,
G.A.Jansen,
R.J.Wanders,
and
M.D.Lloyd
(2003).
The chemical biology of branched-chain lipid metabolism.
|
| |
Prog Lipid Res,
42,
359-376.
|
 |
|
|
|
|
 |
P.N.Black,
and
C.C.DiRusso
(2003).
Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification.
|
| |
Microbiol Mol Biol Rev,
67,
454.
|
 |
|
|
|
|
 |
R.Finking,
A.Neumüller,
J.Solsbacher,
D.Konz,
G.Kretzschmar,
M.Schweitzer,
T.Krumm,
and
M.A.Marahiel
(2003).
Aminoacyl adenylate substrate analogues for the inhibition of adenylation domains of nonribosomal peptide synthetases.
|
| |
Chembiochem,
4,
903-906.
|
 |
|
|
|
|
 |
S.A.Sieber,
and
M.A.Marahiel
(2003).
Learning from nature's drug factories: nonribosomal synthesis of macrocyclic peptides.
|
| |
J Bacteriol,
185,
7036-7043.
|
 |
|
|
|
|
 |
T.Duerfahrt,
S.Doekel,
T.Sonke,
P.J.Quaedflieg,
and
M.A.Marahiel
(2003).
Construction of hybrid peptide synthetases for the production of alpha-l-aspartyl-l-phenylalanine, a precursor for the high-intensity sweetener aspartame.
|
| |
Eur J Biochem,
270,
4555-4563.
|
 |
|
|
|
|
 |
A.R.Horswill,
and
J.C.Escalante-Semerena
(2002).
Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis.
|
| |
Biochemistry,
41,
2379-2387.
|
 |
|
|
|
|
 |
D.Mossialos,
U.Ochsner,
C.Baysse,
P.Chablain,
J.P.Pirnay,
N.Koedam,
H.Budzikiewicz,
D.U.Fernández,
M.Schäfer,
J.Ravel,
and
P.Cornelis
(2002).
Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine.
|
| |
Mol Microbiol,
45,
1673-1685.
|
 |
|
|
|
|
 |
J.D.Weimar,
C.C.DiRusso,
R.Delio,
and
P.N.Black
(2002).
Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. Amino acid residues within the ATP/AMP signature motif of Escherichia coli FadD are required for enzyme activity and fatty acid transport.
|
| |
J Biol Chem,
277,
29369-29376.
|
 |
|
|
|
|
 |
J.J.May,
N.Kessler,
M.A.Marahiel,
and
M.T.Stubbs
(2002).
Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
|
| |
Proc Natl Acad Sci U S A,
99,
12120-12125.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.A.Sieber,
U.Linne,
N.J.Hillson,
E.Roche,
C.T.Walsh,
and
M.A.Marahiel
(2002).
Evidence for a monomeric structure of nonribosomal Peptide synthetases.
|
| |
Chem Biol,
9,
997.
|
 |
|
|
|
|
 |
S.Reverchon,
C.Rouanet,
D.Expert,
and
W.Nasser
(2002).
Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity.
|
| |
J Bacteriol,
184,
654-665.
|
 |
|
|
|
|
 |
S.Smith
(2002).
Modular NRPSs are monomeric.
|
| |
Chem Biol,
9,
955-956.
|
 |
|
|
|
|
 |
T.A.Keating,
C.G.Marshall,
C.T.Walsh,
and
A.E.Keating
(2002).
The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains.
|
| |
Nat Struct Biol,
9,
522-526.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Bergendahl,
U.Linne,
and
M.A.Marahiel
(2002).
Mutational analysis of the C-domain in nonribosomal peptide synthesis.
|
| |
Eur J Biochem,
269,
620-629.
|
 |
|
|
|
|
 |
V.J.Starai,
I.Celic,
R.N.Cole,
J.D.Boeke,
and
J.C.Escalante-Semerena
(2002).
Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine.
|
| |
Science,
298,
2390-2392.
|
 |
|
|
|
|
 |
V.R.Viviani,
A.Uchida,
W.Viviani,
and
Y.Ohmiya
(2002).
The influence of Ala243 (Gly247), Arg215 and Thr226 (Asn230) on the bioluminescence spectra and pH-sensitivity of railroad worm, click beetle and firefly luciferases.
|
| |
Photochem Photobiol,
76,
538-544.
|
 |
|
|
|
|
 |
B.K.Scholz-Schroeder,
M.L.Hutchison,
I.Grgurina,
and
D.C.Gross
(2001).
The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis.
|
| |
Mol Plant Microbe Interact,
14,
336-348.
|
 |
|
|
|
|
 |
B.R.Branchini,
R.A.Magyar,
M.H.Murtiashaw,
and
N.C.Portier
(2001).
The role of active site residue arginine 218 in firefly luciferase bioluminescence.
|
| |
Biochemistry,
40,
2410-2418.
|
 |
|
|
|
|
 |
D.Schwarzer,
H.D.Mootz,
and
M.A.Marahiel
(2001).
Exploring the impact of different thioesterase domains for the design of hybrid peptide synthetases.
|
| |
Chem Biol,
8,
997.
|
 |
|
|
|
|
 |
J.Ehlting,
J.J.Shin,
and
C.J.Douglas
(2001).
Identification of 4-coumarate:coenzyme A ligase (4CL) substrate recognition domains.
|
| |
Plant J,
27,
455-465.
|
 |
|
|
|
|
 |
S.Doekel,
and
M.A.Marahiel
(2001).
Biosynthesis of natural products on modular peptide synthetases.
|
| |
Metab Eng,
3,
64-77.
|
 |
|
|
|
|
 |
W.M.Yuan,
G.D.Gentil,
A.D.Budde,
and
S.A.Leong
(2001).
Characterization of the Ustilago maydis sid2 gene, encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster.
|
| |
J Bacteriol,
183,
4040-4051.
|
 |
|
|
|
|
 |
B.R.Branchini,
M.H.Murtiashaw,
R.A.Magyar,
and
S.M.Anderson
(2000).
The role of lysine 529, a conserved residue of the acyl-adenylate-forming enzyme superfamily, in firefly luciferase.
|
| |
Biochemistry,
39,
5433-5440.
|
 |
|
|
|
|
 |
D.E.Ehmann,
C.A.Shaw-Reid,
H.C.Losey,
and
C.T.Walsh
(2000).
The EntF and EntE adenylation domains of Escherichia coli enterobactin synthetase: sequestration and selectivity in acyl-AMP transfers to thiolation domain cosubstrates.
|
| |
Proc Natl Acad Sci U S A,
97,
2509-2514.
|
 |
|
|
|
|
 |
G.Desogus,
F.Todone,
P.Brick,
and
S.Onesti
(2000).
Active site of lysyl-tRNA synthetase: structural studies of the adenylation reaction.
|
| |
Biochemistry,
39,
8418-8425.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.L.Challis,
J.Ravel,
and
C.A.Townsend
(2000).
Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains.
|
| |
Chem Biol,
7,
211-224.
|
 |
|
|
|
|
 |
H.D.Mootz,
D.Schwarzer,
and
M.A.Marahiel
(2000).
Construction of hybrid peptide synthetases by module and domain fusions.
|
| |
Proc Natl Acad Sci U S A,
97,
5848-5853.
|
 |
|
|
|
|
 |
I.Molnár,
T.Schupp,
M.Ono,
R.Zirkle,
M.Milnamow,
B.Nowak-Thompson,
N.Engel,
C.Toupet,
A.Stratmann,
D.D.Cyr,
J.Gorlach,
J.M.Mayo,
A.Hu,
S.Goff,
J.Schmid,
and
J.M.Ligon
(2000).
The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90.
|
| |
Chem Biol,
7,
97.
|
 |
|
|
|
|
 |
J.W.Jung,
J.H.An,
K.B.Na,
Y.S.Kim,
and
W.Lee
(2000).
The active site and substrates binding mode of malonyl-CoA synthetase determined by transferred nuclear Overhauser effect spectroscopy, site-directed mutagenesis, and comparative modeling studies.
|
| |
Protein Sci,
9,
1294-1303.
|
 |
|
|
|
|
 |
L.Du,
C.Sánchez,
M.Chen,
D.J.Edwards,
and
B.Shen
(2000).
The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase.
|
| |
Chem Biol,
7,
623-642.
|
 |
|
|
|
|
 |
M.C.Moffitt,
and
B.A.Neilan
(2000).
The expansion of mechanistic and organismic diversity associated with non-ribosomal peptides.
|
| |
FEMS Microbiol Lett,
191,
159-167.
|
 |
|
|
|
|
 |
S.Doekel,
and
M.A.Marahiel
(2000).
Dipeptide formation on engineered hybrid peptide synthetases.
|
| |
Chem Biol,
7,
373-384.
|
 |
|
|
|
|
 |
T.Weber,
R.Baumgartner,
C.Renner,
M.A.Marahiel,
and
T.A.Holak
(2000).
Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases.
|
| |
Structure,
8,
407-418.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
U.Linne,
and
M.A.Marahiel
(2000).
Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization.
|
| |
Biochemistry,
39,
10439-10447.
|
 |
|
|
|
|
 |
V.R.Viviani,
and
Y.Ohmiya
(2000).
Bioluminescence color determinants of Phrixothrix railroad-worm luciferases: chimeric luciferases, site-directed mutagenesis of Arg 215 and guanidine effect.
|
| |
Photochem Photobiol,
72,
267-271.
|
 |
|
|
|
|
 |
C.C.DiRusso,
P.N.Black,
and
J.D.Weimar
(1999).
Molecular inroads into the regulation and metabolism of fatty acids, lessons from bacteria.
|
| |
Prog Lipid Res,
38,
129-197.
|
 |
|
|
|
|
 |
C.L.Bender,
F.Alarcón-Chaidez,
and
D.C.Gross
(1999).
Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases.
|
| |
Microbiol Mol Biol Rev,
63,
266-292.
|
 |
|
|
|
|
 |
D.Konz,
and
M.A.Marahiel
(1999).
How do peptide synthetases generate structural diversity?
|
| |
Chem Biol,
6,
R39-R48.
|
 |
|
|
|
|
 |
D.Konz,
S.Doekel,
and
M.A.Marahiel
(1999).
Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin.
|
| |
J Bacteriol,
181,
133-140.
|
 |
|
|
|
|
 |
H.D.Mootz,
and
M.A.Marahiel
(1999).
Design and application of multimodular peptide synthetases.
|
| |
Curr Opin Biotechnol,
10,
341-348.
|
 |
|
|
|
|
 |
K.Reuter,
M.R.Mofid,
M.A.Marahiel,
and
R.Ficner
(1999).
Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
|
| |
EMBO J,
18,
6823-6831.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.J.Belshaw,
C.T.Walsh,
and
T.Stachelhaus
(1999).
Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis.
|
| |
Science,
284,
486-489.
|
 |
|
|
|
|
 |
T.A.Keating,
and
C.T.Walsh
(1999).
Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis.
|
| |
Curr Opin Chem Biol,
3,
598-606.
|
 |
|
|
|
|
 |
T.Stachelhaus,
H.D.Mootz,
and
M.A.Marahiel
(1999).
The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases.
|
| |
Chem Biol,
6,
493-505.
|
 |
|
|
|
|
 |
V.R.Viviani,
E.J.Bechara,
and
Y.Ohmiya
(1999).
Cloning, sequence analysis, and expression of active Phrixothrix railroad-worms luciferases: relationship between bioluminescence spectra and primary structures.
|
| |
Biochemistry,
38,
8271-8279.
|
 |
|
|
|
|
 |
A.M.Gehring,
I.Mori,
R.D.Perry,
and
C.T.Walsh
(1998).
The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis.
|
| |
Biochemistry,
37,
11637-11650.
|
 |
|
|
|
|
 |
B.R.Branchini,
R.A.Magyar,
M.H.Murtiashaw,
S.M.Anderson,
and
M.Zimmer
(1998).
Site-directed mutagenesis of histidine 245 in firefly luciferase: a proposed model of the active site.
|
| |
Biochemistry,
37,
15311-15319.
|
 |
|
|
|
|
 |
D.Sung,
and
H.Kang
(1998).
The N-terminal amino acid sequences of the firefly luciferase are important for the stability of the enzyme.
|
| |
Photochem Photobiol,
68,
749-753.
|
 |
|
|
|
|
 |
E.Guenzi,
G.Galli,
I.Grgurina,
E.Pace,
P.Ferranti,
and
G.Grandi
(1998).
Coordinate transcription and physical linkage of domains in surfactin synthetase are not essential for proper assembly and activity of the multienzyme complex.
|
| |
J Biol Chem,
273,
14403-14410.
|
 |
|
|
|
|
 |
M.Rizzi,
M.Bolognesi,
and
A.Coda
(1998).
A novel deamido-NAD+-binding site revealed by the trapped NAD-adenylate intermediate in the NAD+ synthetase structure.
|
| |
Structure,
6,
1129-1140.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.P.Franks,
A.Jenkins,
E.Conti,
W.R.Lieb,
and
P.Brick
(1998).
Structural basis for the inhibition of firefly luciferase by a general anesthetic.
|
| |
Biophys J,
75,
2205-2211.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.H.Weinreb,
L.E.Quadri,
C.T.Walsh,
and
P.Zuber
(1998).
Stoichiometry and specificity of in vitro phosphopantetheinylation and aminoacylation of the valine-activating module of surfactin synthetase.
|
| |
Biochemistry,
37,
1575-1584.
|
 |
|
|
|
|
 |
R.Herbst,
K.Gast,
and
R.Seckler
(1998).
Folding of firefly (Photinus pyralis) luciferase: aggregation and reactivation of unfolding intermediates.
|
| |
Biochemistry,
37,
6586-6597.
|
 |
|
|
|
|
 |
T.Stachelhaus,
H.D.Mootz,
V.Bergendahl,
and
M.A.Marahiel
(1998).
Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain.
|
| |
J Biol Chem,
273,
22773-22781.
|
 |
|
|
|
|
 |
W.Kallow,
H.von Döhren,
and
H.Kleinkauf
(1998).
Penicillin biosynthesis: energy requirement for tripeptide precursor formation by delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase from Acremonium chrysogenum.
|
| |
Biochemistry,
37,
5947-5952.
|
 |
|
|
|
|
 |
C.A.Townsend
(1997).
Structural studies of natural product biosynthetic proteins.
|
| |
Chem Biol,
4,
721-730.
|
 |
|
|
|
|
 |
C.J.Schofield,
J.E.Baldwin,
M.F.Byford,
I.Clifton,
J.Hajdu,
C.Hensgens,
and
P.Roach
(1997).
Proteins of the penicillin biosynthesis pathway.
|
| |
Curr Opin Struct Biol,
7,
857-864.
|
 |
|
|
|
|
 |
H.D.Mootz,
and
M.A.Marahiel
(1997).
The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains.
|
| |
J Bacteriol,
179,
6843-6850.
|
 |
|
|
|
|
 |
H.D.Mootz,
and
M.A.Marahiel
(1997).
Biosynthetic systems for nonribosomal peptide antibiotic assembly.
|
| |
Curr Opin Chem Biol,
1,
543-551.
|
 |
|
|
|
|
 |
M.A.Marahiel
(1997).
Protein templates for the biosynthesis of peptide antibiotics.
|
| |
Chem Biol,
4,
561-567.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

