 |
PDBsum entry 1alh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase (phosphoric monoester)
|
PDB id
|
|

|

|
1alh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.1
- alkaline phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a phosphate monoester + H2O = an alcohol + phosphate
|
 |
 |
 |
 |
 |
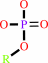 phosphate monoester
phosphate monoester
|
+
|
H2O
|
=
|
 alcohol
alcohol
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+); Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
3:2005-2014
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Kinetics and crystal structure of a mutant Escherichia coli alkaline phosphatase (Asp-369-->Asn): a mechanism involving one zinc per active site.
|
|
T.T.Tibbitts,
X.Xu,
E.R.Kantrowitz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Using site-directed mutagenesis, an aspartate side chain involved in binding
metal ions in the active site of Escherichia coli alkaline phosphatase (Asp-369)
was replaced, alternately, by asparagine (D369N) and by alanine (D369A). The
purified mutant enzymes showed reduced turnover rates (kcat) and increased
Michaelis constants (Km). The kcat for the D369A enzyme was 5,000-fold lower
than the value for the wild-type enzyme. The D369N enzyme required Zn2+ in
millimolar concentrations to become fully active; even under these conditions
the kcat measured for hydrolysis of p-nitrophenol phosphate was 2 orders of
magnitude lower than for the wild-type enzyme. Thus the kcat/Km ratios showed
that catalysis is 50 times less efficient when the carboxylate side chain of
Asp-369 is replaced by the corresponding amide; and activity is reduced to near
nonenzymic levels when the carboxylate is replaced by a methyl group. The
crystal structure of D369N, solved to 2.5 A resolution with an R-factor of
0.189, showed vacancies at 2 of the 3 metal binding sites. On the basis of the
kinetic results and the refined X-ray coordinates, a reaction mechanism is
proposed for phosphate ester hydrolysis by the D369N enzyme involving only 1
metal with the possible assistance of a histidine side chain.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 6.
Fig. 6. Stereo pair showing the electron
density of the anion binding site
ound in D369N alkaline phosphatase.
The ligand at this site, which may be ei-
phosphate or sulfate, was modeled
assulfatedurigthe refinement. Fig-
6,7,and 8 were produced sing the
program SETOR (Evans, 1993).
|
 |
Figure 8.
Fig. 8. pair comparing the posi-
tions of side chains, metals, andphos-
phate in the active sites of the D369N
enzyme (thick lines) and the wild-type
enzyme (thin lines). At the I site, zinc
is bound close to he same location in
both enzymes (crosses), but phosphate
(PO,) is more exposed to the surface in
the D369N structure. At the M2 site, zinc
is bound n the wild-type (cross) but not
theutant enzyme. This permits SI02
to move slightly closer to H370 and the
asparagineintroduce at position 369
(D369N). The M3 site normally contains
Mgz+ (cross) with 3 water molecules
(not shown) in the wild-type enzyme; in
the mutant enzyme this space is partially
filled y 1 water molecule (not shown),
the carboxylate side chain of D5 1, and
the#-aminogroup of K328. The 2 sets of
atomiccordinates were aligned using
Quanta to minimize the RMSD of the Cor
atomsbefore making this omparison.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the Protein Society:
Protein Sci
(1994,
3,
2005-2014)
copyright 1994.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
W.Li,
L.Bi,
W.Wang,
Y.Li,
Y.Zhou,
H.Wei,
T.Jiang,
L.Bai,
Y.Chen,
Z.Zhang,
X.Yuan,
J.Xiao,
and
X.E.Zhang
(2009).
Development of a universal phosphorylated peptide-binding protein for simultaneous assay of kinases.
|
| |
Biosens Bioelectron,
24,
2871-2877.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.H.Muller,
C.Lamoure,
M.H.Le Du,
L.Cattolico,
E.Lajeunesse,
F.Lemaître,
A.Pearson,
F.Ducancel,
A.Ménez,
and
J.C.Boulain
(2001).
Improving Escherichia coli alkaline phosphatase efficacy by additional mutations inside and outside the catalytic pocket.
|
| |
Chembiochem,
2,
517-523.
|
 |
|
|
|
|
 |
Y.D.Park,
Y.Yang,
Q.X.Chen,
H.N.Lin,
Q.Liu,
and
H.M.Zhou
(2001).
Kinetics of complexing activation by the magnesium ion on green crab (Scylla serrata) alkaline phosphatase.
|
| |
Biochem Cell Biol,
79,
765-772.
|
 |
|
|
|
|
 |
M.Bortolato,
F.Besson,
and
B.Roux
(1999).
Role of metal ions on the secondary and quaternary structure of alkaline phosphatase from bovine intestinal mucosa.
|
| |
Proteins,
37,
310-318.
|
 |
|
|
|
|
 |
L.Ma,
T.T.Tibbitts,
and
E.R.Kantrowitz
(1995).
Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
|
| |
Protein Sci,
4,
1498-1506.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

