 |
PDBsum entry 1acm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase (carbamoyl-p,aspartate)
|
PDB id
|
|

|

|
1acm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase (carbamoyl-p,aspartate)
|
 |
|
Title:
|
 |
Arginine 54 in the active site of escherichia coli aspartate transcarbamoylase is critical for catalysis: a site-specific mutagenesis, nmr and x-ray crystallography study
|
|
Structure:
|
 |
Aspartate carbamoyltransferase, catalytic chain. Chain: a, c. Engineered: yes. Aspartate carbamoyltransferase regulatory chain. Chain: b, d. Engineered: yes
|
|
Source:
|
 |
not given
|
|
Biol. unit:
|
 |
 Dodecamer (from
)
Dodecamer (from
)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
R.C.Stevens,E.R.Kantrowitz,W.N.Lipscomb
|
|
Key ref:
|
 |
J.W.Stebbins
et al.
(1992).
Arginine 54 in the active site of Escherichia coli aspartate transcarbamoylase is critical for catalysis: a site-specific mutagenesis, NMR, and X-ray crystallographic study.
Protein Sci,
1,
1435-1446.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
08-Jul-92
|
Release date:
|
15-Jul-92
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, C:
E.C.2.1.3.2
- aspartate carbamoyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
carbamoyl phosphate + L-aspartate = N-carbamoyl-L-aspartate + phosphate + H+
|
 |
 |
 |
 |
 |
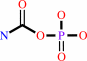 carbamoyl phosphate
carbamoyl phosphate
|
+
|
 L-aspartate
L-aspartate
|
=
|
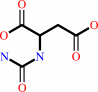 N-carbamoyl-L-aspartate
N-carbamoyl-L-aspartate
|
+
|
phosphate
Bound ligand (Het Group name = )
matches with 64.71% similarity
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains B, D:
E.C.?
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
1:1435-1446
(1992)
|
|
PubMed id:
|
|
|
|

|
| |
|
Arginine 54 in the active site of Escherichia coli aspartate transcarbamoylase is critical for catalysis: a site-specific mutagenesis, NMR, and X-ray crystallographic study.
|
|
J.W.Stebbins,
D.E.Robertson,
M.F.Roberts,
R.C.Stevens,
W.N.Lipscomb,
E.R.Kantrowitz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The replacement of Arg-54 by Ala in the active site of Escherichia coli
aspartate transcarbamoylase causes a 17,000-fold loss of activity but does not
significantly influence the binding of substrates or substrate analogs
(Stebbins, J.W., Xu, W., & Kantrowitz, E.R., 1989, Biochemistry 28,
2592-2600). In the X-ray structure of the wild-type enzyme, Arg-54 interacts
with both the anhydride oxygen and a phosphate oxygen of carbamoyl phosphate
(CP) (Gouaux, J.E. & Lipscomb, W.N., 1988, Proc. Natl. Acad. Sci. USA 85,
4205-4208). The Arg-54-->Ala enzyme was crystallized in the presence of the
transition state analog N-phosphonacetyl-L-aspartate (PALA), data were collected
to a resolution limit of 2.8 A, and the structure was solved by molecular
replacement. The analysis of the refined structure (R factor = 0.18) indicates
that the substitution did not cause any significant alterations to the active
site, except that the side chain of the arginine was replaced by two water
molecules. 31P-NMR studies indicate that the binding of CP to the wild-type
catalytic subunit produces an upfield chemical shift that cannot reflect a
significant change in the ionization state of the CP but rather indicates that
there are perturbations in the electronic environment around the phosphate
moiety when CP binds to the enzyme. The pH dependence of this upfield shift for
bound CP indicates that the catalytic subunit undergoes a conformational change
with a pKa approximately 7.7 upon CP binding. Furthermore, the linewidth of the
31P signal of CP bound to the Arg-54-->Ala enzyme is significantly narrower
than that of CP bound to the wild-type catalytic subunit at any pH, although the
change in chemical shift for the CP bound to the mutant enzyme is unaltered.
31P-NMR studies of PALA complexed to the wild-type catalytic subunit indicate
that the phosphonate group of the bound PALA exists as the dianion at pH 7.0 and
8.8, whereas in the Arg-54-->Ala catalytic subunit the phosphonate group of
the bound PALA exists as the monoanion at pH 7.0 and 8.8. Thus, the side chain
of Arg-54 is essential for the proper ionization of the phosphonate group of
PALA and by analogy the phosphate group in the transition state. These data
support the previously proposed proton transfer mechanism, in which a fully
ionized phosphate group in the transition state accepts a proton during
catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.A.Stieglitz,
J.Xia,
and
E.R.Kantrowitz
(2009).
The first high pH structure of Escherichia coli aspartate transcarbamoylase.
|
| |
Proteins,
74,
318-327.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Helmstaedt,
S.Krappmann,
and
G.H.Braus
(2001).
Allosteric regulation of catalytic activity: Escherichia coli aspartate transcarbamoylase versus yeast chorismate mutase.
|
| |
Microbiol Mol Biol Rev,
65,
404.
|
 |
|
|
|
|
 |
D.Shi,
H.Morizono,
M.Aoyagi,
M.Tuchman,
and
N.M.Allewell
(2000).
Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 A resolution.
|
| |
Proteins,
39,
271-277.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Thomas,
K.Hinsen,
M.J.Field,
and
D.Perahia
(1999).
Tertiary and quaternary conformational changes in aspartate transcarbamylase: a normal mode study.
|
| |
Proteins,
34,
96.
|
 |
|
|
|
|
 |
C.Macol,
M.Dutta,
B.Stec,
H.Tsuruta,
and
E.R.Kantrowitz
(1999).
The 80s loop of the catalytic chain of Escherichia coli aspartate transcarbamoylase is critical for catalysis and homotropic cooperativity.
|
| |
Protein Sci,
8,
1305-1313.
|
 |
|
|
|
|
 |
L.Jin,
B.Stec,
W.N.Lipscomb,
and
E.R.Kantrowitz
(1999).
Insights into the mechanisms of catalysis and heterotropic regulation of Escherichia coli aspartate transcarbamoylase based upon a structure of the enzyme complexed with the bisubstrate analogue N-phosphonacetyl-L-aspartate at 2.1 A.
|
| |
Proteins,
37,
729-742.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.B.Murata,
and
H.K.Schachman
(1996).
Structural similarity between ornithine and aspartate transcarbamoylases of Escherichia coli: characterization of the active site and evidence for an interdomain carboxy-terminal helix in ornithine transcarbamoylase.
|
| |
Protein Sci,
5,
709-718.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

