 |
PDBsum entry 1a43

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Viral protein
|
PDB id
|
|

|

|
1a43
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.7.7.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.7.49
- RNA-directed Dna polymerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
DNA(n) + a 2'-deoxyribonucleoside 5'-triphosphate = DNA(n+1) + diphosphate
|
 |
 |
 |
 |
 |
 DNA(n)
DNA(n)
|
+
|
2'-deoxyribonucleoside 5'-triphosphate
|
=
|
DNA(n+1)
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.7.7.7
- DNA-directed Dna polymerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
DNA(n) + a 2'-deoxyribonucleoside 5'-triphosphate = DNA(n+1) + diphosphate
|
 |
 |
 |
 |
 |
 DNA(n)
DNA(n)
|
+
|
2'-deoxyribonucleoside 5'-triphosphate
|
=
|
DNA(n+1)
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.1.-.-
|
|
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.3.1.13.2
- exoribonuclease H.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Exonucleolytic cleavage to 5'-phosphomonoester oligonucleotides in both 5'- to 3'- and 3'- to 5'-directions.
|
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
E.C.3.1.26.13
- retroviral ribonuclease H.
|
|
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
E.C.3.4.23.16
- HIV-1 retropepsin.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Specific for a P1 residue that is hydrophobic, and P1' variable, but often Pro.
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
55:85-92
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
|
|
D.K.Worthylake,
H.Wang,
S.Yoo,
W.I.Sundquist,
C.P.Hill.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The human immunodeficiency virus type I (HIV-1) capsid protein is initially
synthesized as the central domain of the Gag polyprotein, and is subsequently
proteolytically processed into a discrete 231-amino-acid protein that forms the
distinctive conical core of the mature virus. The crystal structures of two
proteins that span the C-terminal domain of the capsid are reported here: one
encompassing residues 146-231 (CA146-231) and the other extending to include the
14-residue p2 domain of Gag (CA146-p2). The isomorphous CA146-231 and CA146-p2
structures were determined by molecular replacement and have been refined at 2.6
A resolution to R factors of 22.3 and 20.7% (Rfree = 28.1 and 27.5%),
respectively. The ordered domains comprise residues 148-219 for CA146-231 and
148-218 for CA146-p2, and their refined structures are essentially identical.
The proteins are composed of a 310 helix followed by an extended strand and four
alpha-helices. A crystallographic twofold generates a dimer that is stabilized
by parallel packing of an alpha-helix 2 across the dimer interface and by
packing of the 310 helix into a groove created by alpha-helices 2 and 3 of the
partner molecule. CA146-231 and CA146-p2 dimerize with the full affinity of the
intact capsid protein, and their structures therefore reveal the essential dimer
interface of the HIV-1 capsid.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5 View of the CA[146-231]/CA[146-p2] dimer-interface
residues. Residues that appear to lose more than 5 Å^2 of
solvent-accessible surface area upon dimer formation are shown
explicitly. Orientation and color code are the same as for Fig.
3-.
|
 |
Figure 6.
Figure 6 Model of the full-length capsid protein. N-terminal
domains (residues 1-145) are colored magenta and C-terminal
domain dimer (residues 148-219) are colored blue. The disordered
residues Ser146 and Pro147 which link the N- and C-terminal
domains are colored gray. The C-terminal dimer is oriented with
the twofold axis vertical. Atomic models of the full-length
capsid protein were built using the crystal structures of the N-
and C-terminal domains of capsid (Gamble et al., 1996[Gamble, T.
R., Vajdos, F. F., Yoo, S., Worthylake, D. K., Houseweart, M.,
Sundquist, W. I. & Hill, C. P. (1996). Cell, 87, 1285-1294.]).
The last ordered residue of the N-terminal domain (Tyr145) was
covalently connected to the first ordered residue of the
C-terminal domain (Thr148) with a dipeptide linker
(Ser146-Pro147). Plausible conformations for the Ser146-Pro147
linker allowed models to be built that exhibited a wide ( 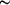 90°)
range of relative rotations between the two domains. 90°)
range of relative rotations between the two domains.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(1999,
55,
85-92)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
 Google scholar
Google scholar
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
O.Pornillos,
B.K.Ganser-Pornillos,
and
M.Yeager
(2011).
Atomic-level modelling of the HIV capsid.
|
| |
Nature,
469,
424-427.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Chen,
and
R.Tycko
(2010).
Structural and dynamical characterization of tubular HIV-1 capsid protein assemblies by solid state nuclear magnetic resonance and electron microscopy.
|
| |
Protein Sci,
19,
716-730.
|
 |
|
|
|
|
 |
C.S.Adamson,
and
E.O.Freed
(2010).
Novel approaches to inhibiting HIV-1 replication.
|
| |
Antiviral Res,
85,
119-141.
|
 |
|
|
|
|
 |
E.B.Monroe,
S.Kang,
S.K.Kyere,
R.Li,
and
P.E.Prevelige
(2010).
Hydrogen/deuterium exchange analysis of HIV-1 capsid assembly and maturation.
|
| |
Structure,
18,
1483-1491.
|
 |
|
|
|
|
 |
J.M.Doolittle,
and
S.M.Gomez
(2010).
Structural similarity-based predictions of protein interactions between HIV-1 and Homo sapiens.
|
| |
Virol J,
7,
82.
|
 |
|
|
|
|
 |
Y.Han,
J.Ahn,
J.Concel,
I.J.Byeon,
A.M.Gronenborn,
J.Yang,
and
T.Polenova
(2010).
Solid-state NMR studies of HIV-1 capsid protein assemblies.
|
| |
J Am Chem Soc,
132,
1976-1987.
|
 |
|
|
|
|
 |
A.M.Gronenborn
(2009).
Protein acrobatics in pairs--dimerization via domain swapping.
|
| |
Curr Opin Struct Biol,
19,
39-49.
|
 |
|
|
|
|
 |
A.P.Mascarenhas,
and
K.Musier-Forsyth
(2009).
The capsid protein of human immunodeficiency virus: interactions of HIV-1 capsid with host protein factors.
|
| |
FEBS J,
276,
6118-6127.
|
 |
|
|
|
|
 |
C.S.Adamson,
K.Salzwedel,
and
E.O.Freed
(2009).
Virus maturation as a new HIV-1 therapeutic target.
|
| |
Expert Opin Ther Targets,
13,
895-908.
|
 |
|
|
|
|
 |
G.Cardone,
J.G.Purdy,
N.Cheng,
R.C.Craven,
and
A.C.Steven
(2009).
Visualization of a missing link in retrovirus capsid assembly.
|
| |
Nature,
457,
694-698.
|
 |
|
|
|
|
 |
G.D.Bailey,
J.K.Hyun,
A.K.Mitra,
and
R.L.Kingston
(2009).
Proton-linked dimerization of a retroviral capsid protein initiates capsid assembly.
|
| |
Structure,
17,
737-748.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.B.Hogue,
A.Hoppe,
and
A.Ono
(2009).
Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding.
|
| |
J Virol,
83,
7322-7336.
|
 |
|
|
|
|
 |
I.J.Byeon,
X.Meng,
J.Jung,
G.Zhao,
R.Yang,
J.Ahn,
J.Shi,
J.Concel,
C.Aiken,
P.Zhang,
and
A.M.Gronenborn
(2009).
Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function.
|
| |
Cell,
139,
780-790.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.G.Purdy,
J.M.Flanagan,
I.J.Ropson,
and
R.C.Craven
(2009).
Retroviral capsid assembly: a role for the CA dimer in initiation.
|
| |
J Mol Biol,
389,
438-451.
|
 |
|
|
|
|
 |
J.L.Neira
(2009).
The capsid protein of human immunodeficiency virus: designing inhibitors of capsid assembly.
|
| |
FEBS J,
276,
6110-6117.
|
 |
|
|
|
|
 |
M.G.Mateu
(2009).
The capsid protein of human immunodeficiency virus: intersubunit interactions during virus assembly.
|
| |
FEBS J,
276,
6098-6109.
|
 |
|
|
|
|
 |
O.Pornillos,
B.K.Ganser-Pornillos,
B.N.Kelly,
Y.Hua,
F.G.Whitby,
C.D.Stout,
W.I.Sundquist,
C.P.Hill,
and
M.Yeager
(2009).
X-ray structures of the hexameric building block of the HIV capsid.
|
| |
Cell,
137,
1282-1292.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.May,
and
M.Zacharias
(2008).
Energy minimization in low-frequency normal modes to efficiently allow for global flexibility during systematic protein-protein docking.
|
| |
Proteins,
70,
794-809.
|
 |
|
|
|
|
 |
B.K.Ganser-Pornillos,
M.Yeager,
and
W.I.Sundquist
(2008).
The structural biology of HIV assembly.
|
| |
Curr Opin Struct Biol,
18,
203-217.
|
 |
|
|
|
|
 |
J.G.Purdy,
J.M.Flanagan,
I.J.Ropson,
K.E.Rennoll-Bankert,
and
R.C.Craven
(2008).
Critical role of conserved hydrophobic residues within the major homology region in mature retroviral capsid assembly.
|
| |
J Virol,
82,
5951-5961.
|
 |
|
|
|
|
 |
L.A.Alcaraz,
M.Del Alamo,
M.G.Mateu,
and
J.L.Neira
(2008).
Structural mobility of the monomeric C-terminal domain of the HIV-1 capsid protein.
|
| |
FEBS J,
275,
3299-3311.
|
 |
|
|
|
|
 |
P.M.Lokhandwala,
T.L.Nguyen,
J.B.Bowzard,
and
R.C.Craven
(2008).
Cooperative role of the MHR and the CA dimerization helix in the maturation of the functional retrovirus capsid.
|
| |
Virology,
376,
191-198.
|
 |
|
|
|
|
 |
P.W.Keller,
M.C.Johnson,
and
V.M.Vogt
(2008).
Mutations in the spacer peptide and adjoining sequences in Rous sarcoma virus Gag lead to tubular budding.
|
| |
J Virol,
82,
6788-6797.
|
 |
|
|
|
|
 |
S.Brun,
M.Solignat,
B.Gay,
E.Bernard,
L.Chaloin,
D.Fenard,
C.Devaux,
N.Chazal,
and
L.Briant
(2008).
VSV-G pseudotyping rescues HIV-1 CA mutations that impair core assembly or stability.
|
| |
Retrovirology,
5,
57.
|
 |
|
|
|
|
 |
V.Bartonova,
S.Igonet,
J.Sticht,
B.Glass,
A.Habermann,
M.C.Vaney,
P.Sehr,
J.Lewis,
F.A.Rey,
and
H.G.Kraüsslich
(2008).
Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
|
| |
J Biol Chem,
283,
32024-32033.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Zhang,
L.Lu,
W.G.Noid,
V.Krishna,
J.Pfaendtner,
and
G.A.Voth
(2008).
A systematic methodology for defining coarse-grained sites in large biomolecules.
|
| |
Biophys J,
95,
5073-5083.
|
 |
|
|
|
|
 |
A.Soutullo,
M.N.Santi,
J.C.Perin,
L.M.Beltramini,
I.M.Borel,
R.Frank,
and
G.G.Tonarelli
(2007).
Systematic epitope analysis of the p26 EIAV core protein.
|
| |
J Mol Recognit,
20,
227-237.
|
 |
|
|
|
|
 |
B.J.Kovaleski,
R.Kennedy,
A.Khorchid,
L.Kleiman,
H.Matsuo,
and
K.Musier-Forsyth
(2007).
Critical role of helix 4 of HIV-1 capsid C-terminal domain in interactions with human lysyl-tRNA synthetase.
|
| |
J Biol Chem,
282,
32274-32279.
|
 |
|
|
|
|
 |
B.K.Ganser-Pornillos,
A.Cheng,
and
M.Yeager
(2007).
Structure of full-length HIV-1 CA: a model for the mature capsid lattice.
|
| |
Cell,
131,
70-79.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.N.Kelly,
S.Kyere,
I.Kinde,
C.Tang,
B.R.Howard,
H.Robinson,
W.I.Sundquist,
M.F.Summers,
and
C.P.Hill
(2007).
Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein.
|
| |
J Mol Biol,
373,
355-366.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.R.Wright,
J.B.Schooler,
H.J.Ding,
C.Kieffer,
C.Fillmore,
W.I.Sundquist,
and
G.J.Jensen
(2007).
Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells.
|
| |
EMBO J,
26,
2218-2226.
|
 |
|
|
|
|
 |
H.Li,
J.Dou,
L.Ding,
and
P.Spearman
(2007).
Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells.
|
| |
J Virol,
81,
12899-12910.
|
 |
|
|
|
|
 |
J.L.Spidel,
C.B.Wilson,
R.C.Craven,
and
J.W.Wills
(2007).
Genetic Studies of the beta-hairpin loop of Rous sarcoma virus capsid protein.
|
| |
J Virol,
81,
1288-1296.
|
 |
|
|
|
|
 |
A.Joshi,
K.Nagashima,
and
E.O.Freed
(2006).
Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation.
|
| |
J Virol,
80,
7939-7951.
|
 |
|
|
|
|
 |
B.J.Kovaleski,
R.Kennedy,
M.K.Hong,
S.A.Datta,
L.Kleiman,
A.Rein,
and
K.Musier-Forsyth
(2006).
In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase.
|
| |
J Biol Chem,
281,
19449-19456.
|
 |
|
|
|
|
 |
C.S.Adamson,
S.D.Ablan,
I.Boeras,
R.Goila-Gaur,
F.Soheilian,
K.Nagashima,
F.Li,
K.Salzwedel,
M.Sakalian,
C.T.Wild,
and
E.O.Freed
(2006).
In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat).
|
| |
J Virol,
80,
10957-10971.
|
 |
|
|
|
|
 |
H.H.Chu,
Y.F.Chang,
and
C.T.Wang
(2006).
Mutations in the alpha-helix directly C-terminal to the major homology region of human immunodeficiency virus type 1 capsid protein disrupt Gag multimerization and markedly impair virus particle production.
|
| |
J Biomed Sci,
13,
645-656.
|
 |
|
|
|
|
 |
K.Oresic,
V.Noriega,
L.Andrews,
and
D.Tortorella
(2006).
A structural determinant of human cytomegalovirus US2 dictates the down-regulation of class I major histocompatibility molecules.
|
| |
J Biol Chem,
281,
19395-19406.
|
 |
|
|
|
|
 |
D.Ivanov,
J.R.Stone,
J.L.Maki,
T.Collins,
and
G.Wagner
(2005).
Mammalian SCAN domain dimer is a domain-swapped homolog of the HIV capsid C-terminal domain.
|
| |
Mol Cell,
17,
137-143.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Ternois,
J.Sticht,
S.Duquerroy,
H.G.Kräusslich,
and
F.A.Rey
(2005).
The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor.
|
| |
Nat Struct Mol Biol,
12,
678-682.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Sticht,
M.Humbert,
S.Findlow,
J.Bodem,
B.Müller,
U.Dietrich,
J.Werner,
and
H.G.Kräusslich
(2005).
A peptide inhibitor of HIV-1 assembly in vitro.
|
| |
Nat Struct Mol Biol,
12,
671-677.
|
 |
|
|
|
|
 |
N.Morellet,
S.Druillennec,
C.Lenoir,
S.Bouaziz,
and
B.P.Roques
(2005).
Helical structure determined by NMR of the HIV-1 (345-392)Gag sequence, surrounding p2: implications for particle assembly and RNA packaging.
|
| |
Protein Sci,
14,
375-386.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Guo,
B.B.Roy,
J.Hu,
A.Roldan,
M.A.Wainberg,
and
C.Liang
(2005).
The R362A mutation at the C-terminus of CA inhibits packaging of human immunodeficiency virus type 1 RNA.
|
| |
Virology,
343,
190-200.
|
 |
|
|
|
|
 |
B.K.Ganser-Pornillos,
U.K.von Schwedler,
K.M.Stray,
C.Aiken,
and
W.I.Sundquist
(2004).
Assembly properties of the human immunodeficiency virus type 1 CA protein.
|
| |
J Virol,
78,
2545-2552.
|
 |
|
|
|
|
 |
J.L.Newman,
E.W.Butcher,
D.T.Patel,
Y.Mikhaylenko,
and
M.F.Summers
(2004).
Flexibility in the P2 domain of the HIV-1 Gag polyprotein.
|
| |
Protein Sci,
13,
2101-2107.
|
 |
|
|
|
|
 |
J.Lanman,
T.T.Lam,
M.R.Emmett,
A.G.Marshall,
M.Sakalian,
and
P.E.Prevelige
(2004).
Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange.
|
| |
Nat Struct Mol Biol,
11,
676-677.
|
 |
|
|
|
|
 |
Y.M.Ma,
and
V.M.Vogt
(2004).
Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro.
|
| |
J Virol,
78,
52-60.
|
 |
|
|
|
|
 |
B.K.Ganser,
A.Cheng,
W.I.Sundquist,
and
M.Yeager
(2003).
Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly.
|
| |
EMBO J,
22,
2886-2892.
|
 |
|
|
|
|
 |
C.Liang,
J.Hu,
J.B.Whitney,
L.Kleiman,
and
M.A.Wainberg
(2003).
A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the gag protein of human immunodeficiency virus type 1.
|
| |
J Virol,
77,
1772-1783.
|
 |
|
|
|
|
 |
J.A.Briggs,
T.Wilk,
R.Welker,
H.G.Kräusslich,
and
S.D.Fuller
(2003).
Structural organization of authentic, mature HIV-1 virions and cores.
|
| |
EMBO J,
22,
1707-1715.
|
 |
|
|
|
|
 |
M.del Alamo,
J.L.Neira,
and
M.G.Mateu
(2003).
Thermodynamic dissection of a low affinity protein-protein interface involved in human immunodeficiency virus assembly.
|
| |
J Biol Chem,
278,
27923-27929.
|
 |
|
|
|
|
 |
S.M.Rue,
J.W.Roos,
L.M.Amzel,
J.E.Clements,
and
S.A.Barber
(2003).
Hydrogen bonding at a conserved threonine in lentivirus capsid is required for virus replication.
|
| |
J Virol,
77,
8009-8018.
|
 |
|
|
|
|
 |
C.Liang,
J.Hu,
R.S.Russell,
A.Roldan,
L.Kleiman,
and
M.A.Wainberg
(2002).
Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag.
|
| |
J Virol,
76,
11729-11737.
|
 |
|
|
|
|
 |
C.Tang,
Y.Ndassa,
and
M.F.Summers
(2002).
Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein.
|
| |
Nat Struct Biol,
9,
537-543.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Fabiola,
R.Bertram,
A.Korostelev,
and
M.S.Chapman
(2002).
An improved hydrogen bond potential: impact on medium resolution protein structures.
|
| |
Protein Sci,
11,
1415-1423.
|
 |
|
|
|
|
 |
J.Lanman,
J.Sexton,
M.Sakalian,
and
P.E.Prevelige
(2002).
Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro.
|
| |
J Virol,
76,
6900-6908.
|
 |
|
|
|
|
 |
M.V.Nermut,
P.Bron,
D.Thomas,
M.Rumlova,
T.Ruml,
and
E.Hunter
(2002).
Molecular organization of Mason-Pfizer monkey virus capsids assembled from Gag polyprotein in Escherichia coli.
|
| |
J Virol,
76,
4321-4330.
|
 |
|
|
|
|
 |
Y.M.Ma,
and
V.M.Vogt
(2002).
Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly.
|
| |
J Virol,
76,
5452-5462.
|
 |
|
|
|
|
 |
T.M.Cairns,
and
R.C.Craven
(2001).
Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants.
|
| |
J Virol,
75,
242-250.
|
 |
|
|
|
|
 |
A.Ono,
D.Demirov,
and
E.O.Freed
(2000).
Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding.
|
| |
J Virol,
74,
5142-5150.
|
 |
|
|
|
|
 |
I.Gross,
H.Hohenberg,
T.Wilk,
K.Wiegers,
M.Grättinger,
B.Müller,
S.Fuller,
and
H.G.Kräusslich
(2000).
A conformational switch controlling HIV-1 morphogenesis.
|
| |
EMBO J,
19,
103-113.
|
 |
|
|
|
|
 |
R.L.Kingston,
T.Fitzon-Ostendorp,
E.Z.Eisenmesser,
G.W.Schatz,
V.M.Vogt,
C.B.Post,
and
M.G.Rossmann
(2000).
Structure and self-association of the Rous sarcoma virus capsid protein.
|
| |
Structure,
8,
617-628.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Monaco-Malbet,
C.Berthet-Colominas,
A.Novelli,
N.Battaï,
N.Piga,
V.Cheynet,
F.Mallet,
and
S.Cusack
(2000).
Mutual conformational adaptations in antigen and antibody upon complex formation between an Fab and HIV-1 capsid protein p24.
|
| |
Structure,
8,
1069-1077.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Morikawa,
D.J.Hockley,
M.V.Nermut,
and
I.M.Jones
(2000).
Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly.
|
| |
J Virol,
74,
16-23.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

