 |
PDBsum entry 1zui

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.71
- shikimate kinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Shikimate and Chorismate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
shikimate + ATP = 3-phosphoshikimate + ADP + H+
|
 |
 |
 |
 |
 |
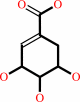 shikimate
shikimate
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
3-phosphoshikimate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Bacteriol
187:8156-8163
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for shikimate-binding specificity of Helicobacter pylori shikimate kinase.
|
|
W.C.Cheng,
Y.N.Chang,
W.C.Wang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Shikimate kinase (EC 2.7.1.71) catalyzes the specific phosphorylation of the
3-hydroxyl group of shikimic acid in the presence of ATP. As the fifth key step
in the shikimate pathway for aromatic amino acid biosynthesis in bacteria,
fungi, and plants, but not mammals, shikimate kinase represents an attractive
target for the development of new antimicrobial agents, herbicides, and
antiparasitic agents. Here, we report the 1.8-Angstroms crystal structure of
Helicobacter pylori shikimate kinase (HpSK). The crystal structure shows a
three-layer alpha/beta fold consisting of a central sheet of five parallel
beta-strands flanked by seven alpha-helices. An HpSK-shikimate-PO(4) complex was
also determined and refined to 2.3 Angstroms, revealing induced-fit movement
from an open to a closed form on substrate binding. Shikimate is located above a
short 3(10) helix formed by a strictly conserved motif (GGGXV) after beta(3).
Moreover, several highly conserved charged residues including Asp33 (in a
conserved DT/SD motif), Arg57, and Arg132 (interacting with shikimate) are
identified, guiding the development of novel inhibitors of shikimate kinase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.J.Duckworth,
A.S.Okoli,
and
G.L.Mendz
(2009).
Novel Helicobacter pylori therapeutic targets: the unusual suspects.
|
| |
Expert Rev Anti Infect Ther,
7,
835-867.
|
 |
|
|
|
|
 |
G.Fucile,
S.Falconer,
and
D.Christendat
(2008).
Evolutionary diversification of plant shikimate kinase gene duplicates.
|
| |
PLoS Genet,
4,
e1000292.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

