 |
PDBsum entry 1yw6

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of succinylglutamate desuccinylase from escherichia coli, northeast structural genomics target et72.
|
|
Structure:
|
 |
Succinylglutamate desuccinylase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: aste. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
3.10Å
|
R-factor:
|
0.227
|
R-free:
|
0.289
|
|
|
Authors:
|
 |
F.Forouhar,W.Yong,A.P.Kuzin,M.Ciano,T.B.Acton,G.T.Montelione,L.Tong, J.F.Hunt,Northeast Structural Genomics Consortium (Nesg)
|
|
Key ref:
|
 |
F.Forouhar
et al.
Crystal structure of succinylglutamate desuccinylase from escherichia coli, Northeast structural genomics target et72..
To be published,
.
|
 |
|
Date:
|
 |
|
17-Feb-05
|
Release date:
|
08-Mar-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P76215
(ASTE_ECOLI) -
Succinylglutamate desuccinylase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
322 a.a.
316 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.96
- succinylglutamate desuccinylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-succinyl-L-glutamate + H2O = L-glutamate + succinate
|
 |
 |
 |
 |
 |
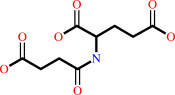 N-succinyl-L-glutamate
N-succinyl-L-glutamate
|
+
|
H2O
|
=
|
 L-glutamate
L-glutamate
|
+
|
 succinate
succinate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cobalt cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

