 |
PDBsum entry 1xm3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
1xm3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Biosynthetic protein
|
 |
|
Title:
|
 |
Crystal structure of northeast structural genomics target sr156
|
|
Structure:
|
 |
Thiazole biosynthesis protein thig. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Gene: thig. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.212
|
R-free:
|
0.237
|
|
|
Authors:
|
 |
A.Kuzin,M.Abashidze,S.Vorobiev,F.Forouhar,T.Acton,L.Ma,R.Xiao, G.Montelione,L.Tong,J.Hunt,Northeast Structural Genomics Consortium (Nesg)
|
|
Key ref:
|
 |
A.Kuzin
et al.
Crystal structure of northeast structural genomics target sr156.
To be published,
.
|
 |
|
Date:
|
 |
|
01-Oct-04
|
Release date:
|
19-Oct-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O31618
(THIG_BACSU) -
Thiazole synthase from Bacillus subtilis (strain 168)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
256 a.a.
244 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.8.1.10
- thiazole synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[ThiS sulfur-carrier protein]-C-terminal-Gly-aminoethanethioate + 2-iminoacetate + 1-deoxy-D-xylulose 5-phosphate = [ThiS sulfur-carrier protein]-C-terminal Gly-Gly + 2-[(2R,5Z)-2-carboxy-4-methylthiazol-5(2H)- ylidene]ethyl phosphate + 2 H2O + H+
|
 |
 |
 |
 |
 |
[ThiS sulfur-carrier protein]-C-terminal-Gly-aminoethanethioate
|
+
|
2-iminoacetate
|
+
|
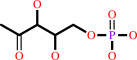 1-deoxy-D-xylulose 5-phosphate
1-deoxy-D-xylulose 5-phosphate
|
=
|
[ThiS sulfur-carrier protein]-C-terminal Gly-Gly
|
+
|
2-[(2R,5Z)-2-carboxy-4-methylthiazol-5(2H)- ylidene]ethyl phosphate
|
+
|
2
×
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

