 |
PDBsum entry 1xix

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.2.10
- UDP-N-acetylmuramoylpentapeptide-lysine N(6)-alanyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
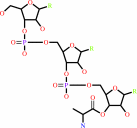
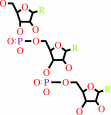
UDP-N-acetyl-alpha-D-muramoyl-L-alanyl-gamma-D-glutamyl-L-lysyl-D-alanyl- D-alanine + L-alanyl-tRNA(Ala) = UDP-N-acetyl-alpha-D-muramoyl-L-alanyl- gamma-D-glutamyl-N6-(L-alanyl)-L-lysyl-D-alanyl-D-alanine + tRNA(Ala) + H(+)
|
 |
 |
 |
 |
 |
UDP-N-acetyl-alpha-D-muramoyl-L-alanyl-gamma-D-glutamyl-L-lysyl-D-alanyl- D-alanine
|
+
|
 L-alanyl-tRNA(Ala)
L-alanyl-tRNA(Ala)
|
=
|
UDP-N-acetyl-alpha-D-muramoyl-L-alanyl- gamma-D-glutamyl-N(6)-(L-alanyl)-L-lysyl-D-alanyl-D-alanine
|
+
|
 tRNA(Ala)
tRNA(Ala)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Bacteriol
187:3833-3838
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure-based site-directed mutagenesis of the UDP-MurNAc-pentapeptide-binding cavity of the FemX alanyl transferase from Weissella viridescens.
|
|
A.P.Maillard,
S.Biarrotte-Sorin,
R.Villet,
S.Mesnage,
A.Bouhss,
W.Sougakoff,
C.Mayer,
M.Arthur.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Weissella viridescens FemX (FemX(Wv)) belongs to the Fem family of nonribosomal
peptidyl transferases that use aminoacyl-tRNA as the amino acid donor to
synthesize the peptide cross-bridge found in the peptidoglycan of many species
of pathogenic gram-positive bacteria. We have recently solved the crystal
structure of FemX(Wv) in complex with the peptidoglycan precursor
UDP-MurNAc-pentapeptide and report here the site-directed mutagenesis of nine
residues located in the binding cavity for this substrate. Two substitutions,
Lys36Met and Arg211Met, depressed FemX(Wv) transferase activity below detectable
levels without affecting protein folding. Analogues of UDP-MurNAc-pentapeptide
lacking the phosphate groups or the C-terminal D-alanyl residues were not
substrates of the enzyme. These results indicate that Lys36 and Arg211
participate in a complex hydrogen bond network that connects the C-terminal
D-Ala residues to the phosphate groups of UDP-MurNAc-pentapeptide and constrains
the substrate in a conformation that is essential for transferase activity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Banerjee,
S.Chen,
K.Dare,
M.Gilreath,
M.Praetorius-Ibba,
M.Raina,
N.M.Reynolds,
T.Rogers,
H.Roy,
S.S.Yadavalli,
and
M.Ibba
(2010).
tRNAs: cellular barcodes for amino acids.
|
| |
FEBS Lett,
584,
387-395.
|
 |
|
|
|
|
 |
A.Bouhss,
B.Al-Dabbagh,
M.Vincent,
B.Odaert,
M.Aumont-Nicaise,
P.Bressolier,
M.Desmadril,
D.Mengin-Lecreulx,
M.C.Urdaci,
and
J.Gallay
(2009).
Specific interactions of clausin, a new lantibiotic, with lipid precursors of the bacterial cell wall.
|
| |
Biophys J,
97,
1390-1397.
|
 |
|
|
|
|
 |
H.Roy
(2009).
Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol.
|
| |
IUBMB Life,
61,
940-953.
|
 |
|
|
|
|
 |
M.Fonvielle,
M.Chemama,
R.Villet,
M.Lecerf,
A.Bouhss,
J.M.Valéry,
M.Ethève-Quelquejeu,
and
M.Arthur
(2009).
Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis.
|
| |
Nucleic Acids Res,
37,
1589-1601.
|
 |
|
|
|
|
 |
A.Bouhss,
A.E.Trunkfield,
T.D.Bugg,
and
D.Mengin-Lecreulx
(2008).
The biosynthesis of peptidoglycan lipid-linked intermediates.
|
| |
FEMS Microbiol Rev,
32,
208-233.
|
 |
|
|
|
|
 |
J.L.Mainardi,
R.Villet,
T.D.Bugg,
C.Mayer,
and
M.Arthur
(2008).
Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria.
|
| |
FEMS Microbiol Rev,
32,
386-408.
|
 |
|
|
|
|
 |
R.P.Garg,
X.L.Qian,
L.B.Alemany,
S.Moran,
and
R.J.Parry
(2008).
Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA.
|
| |
Proc Natl Acad Sci U S A,
105,
6543-6547.
|
 |
|
|
|
|
 |
J.van Heijenoort
(2007).
Lipid intermediates in the biosynthesis of bacterial peptidoglycan.
|
| |
Microbiol Mol Biol Rev,
71,
620-635.
|
 |
|
|
|
|
 |
R.Villet,
M.Fonvielle,
P.Busca,
M.Chemama,
A.P.Maillard,
J.E.Hugonnet,
L.Dubost,
A.Marie,
N.Josseaume,
S.Mesnage,
C.Mayer,
J.M.Valéry,
M.Ethève-Quelquejeu,
and
M.Arthur
(2007).
Idiosyncratic features in tRNAs participating in bacterial cell wall synthesis.
|
| |
Nucleic Acids Res,
35,
6870-6883.
|
 |
|
|
|
|
 |
X.Dong,
M.Kato-Murayama,
T.Muramatsu,
H.Mori,
M.Shirouzu,
Y.Bessho,
and
S.Yokoyama
(2007).
The crystal structure of leucyl/phenylalanyl-tRNA-protein transferase from Escherichia coli.
|
| |
Protein Sci,
16,
528-534.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

