 |
PDBsum entry 1xi2

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1xi2
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.10.5.1
- ribosyldihydronicotinamide dehydrogenase (quinone).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide + a quinone + H+ = beta-nicotinamide D-riboside + a quinol
|
 |
 |
 |
 |
 |
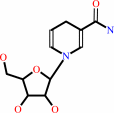 1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide
1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide
|
+
|
 quinone
quinone
|
+
|
H(+)
|
=
|
beta-nicotinamide D-riboside
|
+
|
quinol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Zn(2+)
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
Zn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochem Biophys Res Commun
336:332-338
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of quinone reductase 2 in complex with cancer prodrug CB1954.
|
|
Y.Fu,
L.Buryanovskyy,
Z.Zhang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
CB1954 is a cancer pro-drug that can be activated through reduction by
Escherichia coli nitro-reductases and quinone reductases. Human quinone
reductase 2 is very efficient in the activation of CB1954, approximately 3000
times more efficient than human QR1 in terms of k(cat)/K(m). We have solved the
three-dimensional structure of QR2 in complex with CB1954 to a nominal
resolution of 1.5A. The complex structure indicates the essentiality of the two
nitro groups: one nitro group forms hydrogen bonds with the side-chain of Asn161
of QR2 to hold the other nitro group in position for the reduction. We further
conclude that residue 161, an Asn in QR2 and a His in QR1, is critical in
differentiating the substrate specificities of these two enzymes. Mutation of
Asn161 to His161 in QR2 resulted in the total loss of the enzymatic activity
towards activation of CB1954, whereas the rates of reduction towards menadione
are not altered.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Maiti,
P.V.Reddy,
M.Sturdy,
L.Marler,
S.D.Pegan,
A.D.Mesecar,
J.M.Pezzuto,
and
M.Cushman
(2009).
Synthesis of casimiroin and optimization of its quinone reductase 2 and aromatase inhibitory activities.
|
| |
J Med Chem,
52,
1873-1884.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Winger,
O.Hantschel,
G.Superti-Furga,
and
J.Kuriyan
(2009).
The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2).
|
| |
BMC Struct Biol,
9,
7.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Fu,
L.Buryanovskyy,
and
Z.Zhang
(2008).
Quinone reductase 2 is a catechol quinone reductase.
|
| |
J Biol Chem,
283,
23829-23835.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

