 |
PDBsum entry 1wsv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of human t-protein of glycine cleavage system
|
|
Structure:
|
 |
Aminomethyltransferase. Chain: a, b. Synonym: glycine cleavage system t protein, gcvt. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: gcst. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.171
|
R-free:
|
0.245
|
|
|
Authors:
|
 |
K.Okamura-Ikeda,H.Hosaka,M.Yoshimura,E.Yamashita,S.Toma,A.Nakagawa, K.Fujiwara,Y.Motokawa,H.Taniguchi
|
Key ref:

|
 |
K.Okamura-Ikeda
et al.
(2005).
Crystal structure of human T-protein of glycine cleavage system at 2.0 A resolution and its implication for understanding non-ketotic hyperglycinemia.
J Mol Biol,
351,
1146-1159.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
11-Nov-04
|
Release date:
|
16-Aug-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P48728
(GCST_HUMAN) -
Aminomethyltransferase, mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
403 a.a.
371 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.2.10
- aminomethyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Glycine Cleavage System
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-[(R)-S(8)-aminomethyldihydrolipoyl]-L-lysyl-[protein] + (6S)- 5,6,7,8-tetrahydrofolate = N6-[(R)-dihydrolipoyl]-L-lysyl-[protein] + (6R)-5,10-methylene-5,6,7,8-tetrahydrofolate + NH4+
|
 |
 |
 |
 |
 |
[Protein]-S(8)-aminomethyldihydrolipoyllysine
|
+
|
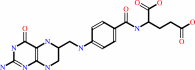 tetrahydrofolate
tetrahydrofolate
|
=
|
[protein]-dihydrolipoyllysine
|
+
|
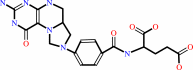 5,10-methylenetetrahydrofolate
5,10-methylenetetrahydrofolate
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
351:1146-1159
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human T-protein of glycine cleavage system at 2.0 A resolution and its implication for understanding non-ketotic hyperglycinemia.
|
|
K.Okamura-Ikeda,
H.Hosaka,
M.Yoshimura,
E.Yamashita,
S.Toma,
A.Nakagawa,
K.Fujiwara,
Y.Motokawa,
H.Taniguchi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
T-protein, a component of the glycine cleavage system, catalyzes the formation
of ammonia and 5,10-methylenetetrahydrofolate from the aminomethyl moiety of
glycine attached to the lipoate cofactor of H-protein. Several mutations in the
human T-protein gene cause non-ketotic hyperglycinemia. To gain insights into
the effect of disease-causing mutations and the catalytic mechanism at the
molecular level, crystal structures of human T-protein in free form and that
bound to 5-methyltetrahydrofolate (5-CH3-H4folate) have been determined at 2.0 A
and 2.6 A resolution, respectively. The overall structure consists of three
domains arranged in a cloverleaf-like structure with the central cavity, where
5-CH3-H4folate is bound in a kinked shape with the pteridine group deeply buried
into the hydrophobic pocket and the glutamyl group pointed to the C-terminal
side surface. Most of the disease-related residues cluster around the cavity,
forming extensive hydrogen bonding networks. These hydrogen bonding networks are
employed in holding not only the folate-binding space but also the positions and
the orientations of alpha-helix G and the following loop in the middle region,
which seems to play a pivotal role in the T-protein catalysis. Structural and
mutational analyses demonstrated that Arg292 interacts through water molecules
with the folate polyglutamate tail, and that the invariant Asp101, located close
to the N10 group of 5-CH3-H4folate, might play a key role in the initiation of
the catalysis by increasing the nucleophilic character of the N10 atom of the
folate substrate for the nucleophilic attack on the aminomethyl lipoate
intermediate. A clever mechanism of recruiting the aminomethyl lipoate arm to
the reaction site seems to function as a way of avoiding the release of toxic
formaldehyde.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Schematic representation of the proposed
aminomethyltransfer reaction catalyzed by T-protein. The bound
H[4]folate attacks the methylene carbon of the aminomethyl
lipoate arm attached to H-protein via the nucleophilic N10 group
putatively with concomitant proton abstraction by Asp101 and
ammonia release. The resulting covalent intermediate is attacked
by the N5 group with concomitant deprotonation of the N5 by the
sulfur group of the nascent dihydrolipoate. An alternative
possibility is that the N5 group attacks first, followed by the
attack of the N10 group.
|
 |
Figure 5.
Figure 5. (a) NKH-related mutation sites mapped on the
overall topology of huT. The mutant residues are depicted in a
ball-and-stick representation with atoms colored in red. Residue
numbers are labeled. (b) Stereo view of the hydrogen bonding
networks as well as the putative functional residues in the huT
reaction site. The side-chains of the NKH-related residues and
residues hydrogen bonded to these are colored in red and green,
respectively. The side-chains of the putative functional
residues are colored in blue. Atoms, ligand and hydrogen bonds
are colored as in Figure 1(b).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2005,
351,
1146-1159)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.S.Tibbetts,
and
D.R.Appling
(2010).
Compartmentalization of Mammalian folate-mediated one-carbon metabolism.
|
| |
Annu Rev Nutr,
30,
57-81.
|
 |
|
|
|
|
 |
G.Kikuchi,
Y.Motokawa,
T.Yoshida,
and
K.Hiraga
(2008).
Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia.
|
| |
Proc Jpn Acad Ser B Phys Biol Sci,
84,
246-263.
|
 |
|
|
|
|
 |
T.Nakai,
S.Kuramitsu,
and
N.Kamiya
(2008).
Structural bases for the specific interactions between the E2 and E3 components of the Thermus thermophilus 2-oxo acid dehydrogenase complexes.
|
| |
J Biochem,
143,
747-758.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

