
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 174 a.a.
174 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 0 more)
160 a.a.
(+ 0 more)
160 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of a type ii thymidine kinase with bound dttp
|
|
Structure:
|
 |
Thymidine kinase. Chain: a, b, c, d, e, f, g, h. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Tetramer (from PDB file)
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
1.83Å
|
R-factor:
|
0.160
|
R-free:
|
0.189
|
|
|
Authors:
|
 |
M.S.Birringer,M.T.Claus,G.Folkers,D.P.Kloer,G.E.Schulz,L.Scapozza
|
Key ref:

|
 |
M.S.Birringer
et al.
(2005).
Structure of a type II thymidine kinase with bound dTTP.
FEBS Lett,
579,
1376-1382.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
27-Jul-04
|
Release date:
|
01-Feb-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.2.7.1.21
- thymidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
thymidine + ATP = dTMP + ADP + H+
|
 |
 |
 |
 |
 |
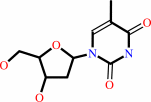 thymidine
thymidine
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 76.47% similarity
|
=
|
 dTMP
dTMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
FEBS Lett
579:1376-1382
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of a type II thymidine kinase with bound dTTP.
|
|
M.S.Birringer,
M.T.Claus,
G.Folkers,
D.P.Kloer,
G.E.Schulz,
L.Scapozza.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of human cytosolic thymidine kinase in complex with its feedback
inhibitor 2'-deoxythymidine-5'-triphosphate was determined. This structure is
the first representative of the type II thymidine kinases found in several
pathogens. The structure deviates strongly from the known structures of type I
thymidine kinases such as the Herpes simplex enzyme. It contains a zinc-binding
domain with four cysteines complexing a structural zinc ion. Interestingly, the
backbone atoms of the type II enzyme bind thymine via hydrogen-bonds, in
contrast to type I, where side chains are involved. This results in a
specificity difference exploited for antiviral therapy. The presented structure
will foster the development of new drugs and prodrugs for numerous therapeutic
applications.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Stereoview of the active center of hTK1. Water
molecules are red balls, hydrogen bonds are dashed lines. (A)
Bound feedback inhibitor dTTP with hydrogen bonding network. The
conformation of dTTP in its two binding modes was derived from
the F[o]–F[c] omit electron density map here contoured at 3σ
(green). The 40% binding mode is shown in a semi transparent
mode. (B) The nucleotide is bound by the tight interactions with
protein residues. Hydrogen bonds to main chain atoms and
stacking of the pyrimidine ring between Phe101, Phe133 and
Tyr181 are responsible for the high substrate specificity.
|
 |
Figure 4.
Fig. 4. B-factor plot of the eight subunits of the reported
hTK1 structure. The map correlation is depicted on the right
side of the diagram. High mobility of some residues leads to
lacking parts in the structure. The line on top shows the
secondary structure assignment as given by the program DSSP
[20]. The β-sheets and helices are displayed as arrows and
tubes, respectively and labeled.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Federation of European Biochemical Societies:
FEBS Lett
(2005,
579,
1376-1382)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.M.Doolittle,
and
S.M.Gomez
(2011).
Mapping Protein Interactions between Dengue Virus and Its Human and Insect Hosts.
|
| |
PLoS Negl Trop Dis,
5,
e954.
|
 |
|
|
|
|
 |
B.Munch-Petersen
(2009).
Reversible tetramerization of human TK1 to the high catalytic efficient form is induced by pyrophosphate, in addition to tripolyphosphates, or high enzyme concentration.
|
| |
FEBS J,
276,
571-580.
|
 |
|
|
|
|
 |
P.Lupieri,
C.H.Nguyen,
Z.G.Bafghi,
A.Giorgetti,
and
P.Carloni
(2009).
Computational molecular biology approaches to ligand-target interactions.
|
| |
HFSP J,
3,
228-239.
|
 |
|
|
|
|
 |
N.E.Mikkelsen,
B.Munch-Petersen,
and
H.Eklund
(2008).
Structural studies of nucleoside analog and feedback inhibitor binding to Drosophila melanogaster multisubstrate deoxyribonucleoside kinase.
|
| |
FEBS J,
275,
2151-2160.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Segura-Peña,
J.Lichter,
M.Trani,
M.Konrad,
A.Lavie,
and
S.Lutz
(2007).
Quaternary structure change as a mechanism for the regulation of thymidine kinase 1-like enzymes.
|
| |
Structure,
15,
1555-1566.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Egeblad-Welin,
Y.Sonntag,
H.Eklund,
and
B.Munch-Petersen
(2007).
Functional studies of active-site mutants from Drosophila melanogaster deoxyribonucleoside kinase. Investigations of the putative catalytic glutamate-arginine pair and of residues responsible for substrate specificity.
|
| |
FEBS J,
274,
1542-1551.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
U.Kosinska,
C.Carnrot,
M.P.Sandrini,
A.R.Clausen,
L.Wang,
J.Piskur,
S.Eriksson,
and
H.Eklund
(2007).
Structural studies of thymidine kinases from Bacillus anthracis and Bacillus cereus provide insights into quaternary structure and conformational changes upon substrate binding.
|
| |
FEBS J,
274,
727-737.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Tjarks,
R.Tiwari,
Y.Byun,
S.Narayanasamy,
and
R.F.Barth
(2007).
Carboranyl thymidine analogues for neutron capture therapy.
|
| |
Chem Commun (Camb),
(),
4978-4991.
|
 |
|
|
|
|
 |
A.Johayem,
S.Raić-Malić,
K.Lazzati,
P.A.Schubiger,
L.Scapozza,
and
S.M.Ametamey
(2006).
Synthesis and characterization of a C6 nucleoside analogue for the in vivo imaging of the gene expression of herpes simplex virus type-1 thymidine kinase (HSV1 TK).
|
| |
Chem Biodivers,
3,
274-283.
|
 |
|
|
|
|
 |
K.El Omari,
N.Solaroli,
A.Karlsson,
J.Balzarini,
and
D.K.Stammers
(2006).
Structure of vaccinia virus thymidine kinase in complex with dTTP: insights for drug design.
|
| |
BMC Struct Biol,
6,
22.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
U.Kosinska,
C.Carnrot,
S.Eriksson,
L.Wang,
and
H.Eklund
(2005).
Structure of the substrate complex of thymidine kinase from Ureaplasma urealyticum and investigations of possible drug targets for the enzyme.
|
| |
FEBS J,
272,
6365-6372.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

