 |
PDBsum entry 1tip

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
The bisphosphatase domain of the bifunctional rat liver 6- phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
Structure:
|
 |
Phosphoenzyme intermediate of fru-2,6-bisphosphatase. Chain: a, b. Synonym: d-fructose-2,6-bisphosphate 2-phosphohydrolase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Rattus norvegicus. Norway rat. Organism_taxid: 10116. Organ: liver. Gene: a coding region which covers. Expressed in: escherichia coli. Expression_system_taxid: 562. Bisphosphatase domain (residues 251 - 440) of the rat liver 6-pf-2- k/fru-2,6-p2ase (residues 1 - 470)
|
|
Biol. unit:
|
 |
Homo-Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.204
|
R-free:
|
0.286
|
|
|
Authors:
|
 |
Y.-H.Lee,T.W.Olson,C.M.Ogata,D.G.Levitt,L.J.Banaszak,A.J.Lange
|
|
Key ref:
|
 |
Y.H.Lee
et al.
(1997).
Crystal structure of a trapped phosphoenzyme during a catalytic reaction.
Nat Struct Biol,
4,
615-618.
PubMed id:
|
 |
|
Date:
|
 |
|
28-May-97
|
Release date:
|
28-Jan-98
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07953
(F261_RAT) -
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 1 from Rattus norvegicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
471 a.a.
191 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.7.1.105
- 6-phosphofructo-2-kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 6-phosphate + ATP = beta-D-fructose 2,6-bisphosphate + ADP + H+
|
 |
 |
 |
 |
 |
beta-D-fructose 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
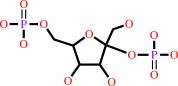 beta-D-fructose 2,6-bisphosphate
beta-D-fructose 2,6-bisphosphate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.46
- fructose-2,6-bisphosphate 2-phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 2,6-bisphosphate + H2O = beta-D-fructose 6-phosphate + phosphate
|
 |
 |
 |
 |
 |
 beta-D-fructose 2,6-bisphosphate
beta-D-fructose 2,6-bisphosphate
|
+
|
H2O
|
=
|
beta-D-fructose 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Struct Biol
4:615-618
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a trapped phosphoenzyme during a catalytic reaction.
|
|
Y.H.Lee,
T.W.Olson,
C.M.Ogata,
D.G.Levitt,
L.J.Banaszak,
A.J.Lange.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the fructose-2,6-bisphosphatase domain trapped during
the reaction reveal a phosphorylated His 258, and a water molecule immobilized
by the product, fructose-6-phosphate. The geometry suggests that the
dephosphorylation step requires prior removal of the product for an 'associative
in-line' phosphoryl transfer to the catalytic water.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Davies,
I.P.Anderson,
P.C.Turner,
A.D.Shirras,
H.H.Rees,
and
D.J.Rigden
(2007).
An unsuspected ecdysteroid/steroid phosphatase activity in the key T-cell regulator, Sts-1: surprising relationship to insect ecdysteroid phosphate phosphatase.
|
| |
Proteins,
67,
720-731.
|
 |
|
|
|
|
 |
H.A.Watkins,
and
E.N.Baker
(2006).
Structural and functional analysis of Rv3214 from Mycobacterium tuberculosis, a protein with conflicting functional annotations, leads to its characterization as a phosphatase.
|
| |
J Bacteriol,
188,
3589-3599.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.G.Kim,
N.P.Manes,
M.R.El-Maghrabi,
and
Y.H.Lee
(2006).
Crystal structure of the hypoxia-inducible form of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3): a possible new target for cancer therapy.
|
| |
J Biol Chem,
281,
2939-2944.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Chevalier,
L.Bertrand,
M.H.Rider,
F.R.Opperdoes,
D.J.Rigden,
and
P.A.Michels
(2005).
6-Phosphofructo-2-kinase and fructose-2,6-bisphosphatase in Trypanosomatidae. Molecular characterization, database searches, modelling studies and evolutionary analysis.
|
| |
FEBS J,
272,
3542-3560.
|
 |
|
|
|
|
 |
Y.H.Lee,
Y.Li,
K.Uyeda,
and
C.A.Hasemann
(2003).
Tissue-specific structure/function differentiation of the liver isoform of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase.
|
| |
J Biol Chem,
278,
523-530.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.A.Okar,
A.Manzano,
A.Navarro-Sabatè,
L.Riera,
R.Bartrons,
and
A.J.Lange
(2001).
PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate.
|
| |
Trends Biochem Sci,
26,
30-35.
|
 |
|
|
|
|
 |
V.Monedero,
S.Poncet,
I.Mijakovic,
S.Fieulaine,
V.Dossonnet,
I.Martin-Verstraete,
S.Nessler,
and
J.Deutscher
(2001).
Mutations lowering the phosphatase activity of HPr kinase/phosphatase switch off carbon metabolism.
|
| |
EMBO J,
20,
3928-3937.
|
 |
|
|
|
|
 |
D.A.Okar,
D.H.Live,
M.H.Devany,
and
A.J.Lange
(2000).
Mechanism of the bisphosphatase reaction of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase probed by (1)H-(15)N NMR spectroscopy.
|
| |
Biochemistry,
39,
9754-9762.
|
 |
|
|
|
|
 |
J.Nairn,
D.Duncan,
N.E.Price,
S.M.Kelly,
L.A.Fothergill-Gilmore,
S.Uhrinova,
P.N.Barlow,
D.J.Rigden,
and
N.C.Price
(2000).
Characterization of active-site mutants of Schizosaccharomyces pombe phosphoglycerate mutase. Elucidation of the roles of amino acids involved in substrate binding and catalysis.
|
| |
Eur J Biochem,
267,
7065-7074.
|
 |
|
|
|
|
 |
D.A.Okar,
and
A.J.Lange
(1999).
Fructose-2,6-bisphosphate and control of carbohydrate metabolism in eukaryotes.
|
| |
Biofactors,
10,
1.
|
 |
|
|
|
|
 |
M.H.Yuen,
H.Mizuguchi,
Y.H.Lee,
P.F.Cook,
K.Uyeda,
and
C.A.Hasemann
(1999).
Crystal structure of the H256A mutant of rat testis fructose-6-phosphate,2-kinase/fructose-2,6-bisphosphatase. Fructose 6-phosphate in the active site leads to mechanisms for both mutant and wild type bisphosphatase activities.
|
| |
J Biol Chem,
274,
2176-2184.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.L.Stoddard
(1998).
New results using Laue diffraction and time-resolved crystallography.
|
| |
Curr Opin Struct Biol,
8,
612-618.
|
 |
|
|
|
|
 |
H.Käck,
K.J.Gibson,
Y.Lindqvist,
and
G.Schneider
(1998).
Snapshot of a phosphorylated substrate intermediate by kinetic crystallography.
|
| |
Proc Natl Acad Sci U S A,
95,
5495-5500.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

