 |
PDBsum entry 1sjb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase, isomerase
|
PDB id
|
|

|

|
1sjb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase, isomerase
|
 |
|
Title:
|
 |
X-ray structure of o-succinylbenzoate synthase complexed with o- succinylbenzoic acid
|
|
Structure:
|
 |
N-acylamino acid racemase. Chain: a, b, c, d. Synonym: o-succinylbenzoate synthase. Engineered: yes
|
|
Source:
|
 |
Amycolatopsis sp.. Organism_taxid: 37632. Gene: aaar. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693.
|
|
Biol. unit:
|
 |
 Octamer (from PDB file)
Octamer (from PDB file)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.209
|
R-free:
|
0.272
|
|
|
Authors:
|
 |
J.B.Thoden,E.A.Taylor-Ringia,J.B.Garrett,J.A.Gerlt,H.M.Holden, I.Rayment
|
Key ref:

|
 |
J.B.Thoden
et al.
(2004).
Evolution of enzymatic activity in the enolase superfamily: structural studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis.
Biochemistry,
43,
5716-5727.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
03-Mar-04
|
Release date:
|
01-Jun-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q44244
(NSAR_AMYSP) -
N-succinylamino acid racemase from Amycolatopsis sp
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
368 a.a.
367 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.2.1.113
- o-succinylbenzoate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1R,6R)-6-hydroxy-2-succinyl-cyclohexa-2,4-diene-1-carboxylate = 2-succinylbenzoate + H2O
|
 |
 |
 |
 |
 |
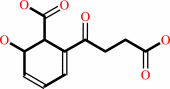 (1R,6R)-6-hydroxy-2-succinyl-cyclohexa-2,4-diene-1-carboxylate
(1R,6R)-6-hydroxy-2-succinyl-cyclohexa-2,4-diene-1-carboxylate
|
=
|
2-succinylbenzoate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+) or Mg(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.1.1.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
43:5716-5727
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Evolution of enzymatic activity in the enolase superfamily: structural studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis.
|
|
J.B.Thoden,
E.A.Taylor Ringia,
J.B.Garrett,
J.A.Gerlt,
H.M.Holden,
I.Rayment.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Divergent evolution of enzyme function is commonly explained by a gene
duplication event followed by mutational changes that allow the protein encoded
by the copy to acquire a new function. An alternate hypothesis is that this
process is facilitated when the progenitor enzyme acquires a second function
while maintaining the original activity. This phenomenon has been suggested to
occur in the o-succinylbenzoate synthase (OSBS) from a species of Amycolatopsis
that catalyzes not only the physiological syn-dehydration reaction of
2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate but also an accidental
racemization of N-acylamino acids [Palmer, D. R., Garrett, J. B., Sharma, V.,
Meganathan, R., Babbitt, P. C., and Gerlt, J. A. (1999) Biochemistry 38,
4252-4258]. To understand the molecular basis of this promiscuity,
three-dimensional structures of liganded complexes of this enzyme have been
determined, including the product of the OSBS reaction and three N-acylamino
acid substrates for the N-acylamino acid racemase (NAAAR) reaction,
N-acetylmethionine, N-succinylmethionine, and N-succinylphenylglycine, to 2.2,
2.3, 2.1, and 1.9 A resolution, respectively. These structures show how the
active-site cavity can accommodate both the hydrophobic substrate for the OSBS
reaction and the substrates for the accidental NAAAR reaction. As expected, the
N-acylamino acid is sandwiched between lysines 163 and 263, which function as
the catalytic bases for the abstraction of the alpha-proton in the (R)- and
(S)-racemization reactions, respectively [Taylor Ringia, E. A., Garrett, J. B,
Thoden, J. B., Holden, H. M., Rayment, I., and Gerlt, J. A. (2004) Biochemistry
42, 224-229]. Importantly, the protein forms specific favorable interactions
with the hydrophobic amino acid side chain, alpha-carbon, carboxylate, and the
polar components of the N-acyl linkage. Accommodation of the components of the
N-acyl linkage appears to be the reason that this enzyme is capable of a
racemization reaction on these substrates, whereas the orthologous OSBS from
Escherichia coli lacks this functionality.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Sakai,
A.A.Fedorov,
E.V.Fedorov,
A.M.Schnoes,
M.E.Glasner,
S.Brown,
M.E.Rutter,
K.Bain,
S.Chang,
T.Gheyi,
J.M.Sauder,
S.K.Burley,
P.C.Babbitt,
S.C.Almo,
and
J.A.Gerlt
(2009).
Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes.
|
| |
Biochemistry,
48,
1445-1453.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
U.Pieper,
R.Chiang,
J.J.Seffernick,
S.D.Brown,
M.E.Glasner,
L.Kelly,
N.Eswar,
J.M.Sauder,
J.B.Bonanno,
S.Swaminathan,
S.K.Burley,
X.Zheng,
M.R.Chance,
S.C.Almo,
J.A.Gerlt,
F.M.Raushel,
M.P.Jacobson,
P.C.Babbitt,
and
A.Sali
(2009).
Target selection and annotation for the structural genomics of the amidohydrolase and enolase superfamilies.
|
| |
J Struct Funct Genomics,
10,
107-125.
|
 |
|
|
|
|
 |
M.Hayashida,
S.H.Kim,
K.Takeda,
T.Hisano,
and
K.Miki
(2008).
Crystal structure of N-acylamino acid racemase from Thermus thermophilus HB8.
|
| |
Proteins,
71,
519-523.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Hult,
and
P.Berglund
(2007).
Enzyme promiscuity: mechanism and applications.
|
| |
Trends Biotechnol,
25,
231-238.
|
 |
|
|
|
|
 |
L.Hou,
M.T.Honaker,
L.M.Shireman,
L.M.Balogh,
A.G.Roberts,
K.C.Ng,
A.Nath,
and
W.M.Atkins
(2007).
Functional promiscuity correlates with conformational heterogeneity in A-class glutathione S-transferases.
|
| |
J Biol Chem,
282,
23264-23274.
|
 |
|
|
|
|
 |
L.Song,
C.Kalyanaraman,
A.A.Fedorov,
E.V.Fedorov,
M.E.Glasner,
S.Brown,
H.J.Imker,
P.C.Babbitt,
S.C.Almo,
M.P.Jacobson,
and
J.A.Gerlt
(2007).
Prediction and assignment of function for a divergent N-succinyl amino acid racemase.
|
| |
Nat Chem Biol,
3,
486-491.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Wroe,
H.S.Chan,
and
E.Bornberg-Bauer
(2007).
A structural model of latent evolutionary potentials underlying neutral networks in proteins.
|
| |
HFSP J,
1,
79-87.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

