 |
PDBsum entry 1qxr

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of phosphoglucose isomerase from pyrococcus furiosus in complex with 5-phosphoarabinonate
|
|
Structure:
|
 |
Glucose-6-phosphate isomerase. Chain: a, b. Synonym: gpi, phosphoglucose isomerase, pgi, phosphohexose isomerase, phi. Engineered: yes
|
|
Source:
|
 |
Pyrococcus furiosus. Organism_taxid: 2261. Gene: pgia or pf0196. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.223
|
R-free:
|
0.264
|
|
|
Authors:
|
 |
M.K.Swan,J.T.G.Solomons,C.C.Beeson,P.Hansen,P.Schonheit,C.Davies
|
Key ref:

|
 |
M.K.Swan
et al.
(2003).
Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus.
J Biol Chem,
278,
47261-47268.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
08-Sep-03
|
Release date:
|
09-Dec-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P83194
(G6PI_PYRFU) -
Glucose-6-phosphate isomerase from Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
189 a.a.
187 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.1.9
- glucose-6-phosphate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-glucose 6-phosphate = beta-D-fructose 6-phosphate
|
 |
 |
 |
 |
 |
alpha-D-glucose 6-phosphate
Bound ligand (Het Group name = )
matches with 82.35% similarity
|
=
|
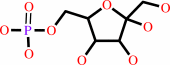 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:47261-47268
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus.
|
|
M.K.Swan,
J.T.Solomons,
C.C.Beeson,
T.Hansen,
P.Schönheit,
C.Davies.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In the Euryarchaeota species Pyrococcus furiosus and Thermococcus litoralis,
phosphoglucose isomerase (PGI) activity is catalyzed by an enzyme unrelated to
the well known family of PGI enzymes found in prokaryotes, eukaryotes, and some
archaea. We have determined the crystal structure of PGI from Pyrococcus
furiosus in native form and in complex with two active site ligands,
5-phosphoarabinonate and gluconate 6-phosphate. In these structures, the metal
ion, which in vivo is presumed to be Fe2+, is located in the core of the cupin
fold and is immediately adjacent to the C1-C2 region of the ligands, suggesting
that Fe2+ is involved in catalysis rather than serving a structural role. The
active site contains a glutamate residue that contacts the substrate, but,
because it is also coordinated to the metal ion, it is highly unlikely to
mediate proton transfer in a cis-enediol mechanism. Consequently, we propose a
hydride shift mechanism of catalysis. In this mechanism, Fe2+ is responsible for
proton transfer between O1 and O2, and the hydride shift between C1 and C2 is
favored by a markedly hydrophobic environment in the active site. The absence of
any obvious enzymatic machinery for catalyzing ring opening of the sugar
substrates suggests that pyrococcal PGI has a preference for straight chain
substrates and that metabolism in extreme thermophiles may use sugars in both
ring and straight chain forms.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIG. 4. The structure of the PfPGI in complex with PAB at
1.7 Å resolution. a, stereo view showing PAB bound to the
active site region of PfPGI. Monomer B of the dimer is shown,
but the contacts are essentially the same in monomer A. The
electron density is a F[o] - F[c] difference map calculated from
the final coordinates refined in the absence of ligand and thus
represents unbiased density of PAB. The side chains of those
residues surrounding the ligand are shown in ball-and-stick form
in which carbons are yellow, oxygens are red, and nitrogens are
blue. The metal ion atom, denoted M, is shown as an orange
sphere, and water molecules are shown as red spheres. Potential
hydrogen bonding and coordination contacts are shown as dashed
lines. The figure was produced using PyMOL (www.pymol.org) (32).
b, diagram of the contacts made between PAB and the enzyme in
which the distances are shown in Ångstroms. The inhibitor
is colored green.
|
 |
Figure 7.
FIG. 7. A catalytic mechanism for phosphoglucose isomerase
from P. furiosus, shown here in the glucose 6-phosphate to
fructose 6-phosphate direction. The substrate binds as the
straight form of G6P, and O1 and O2 displace both water
molecules from the coordination shell around Fe^2+. By
withdrawing electron density from O2, Fe^2+ facilitates the
movement of a proton from O2 to O1, creating a carbocation at
C1. An atom of hydrogen in the form of a hydride then shifts
from C2 to C1. A lone pair of electrons from O2 moves to form a
double bond between O2 and C2, thus creating F6P. When the
product leaves the active site, water molecules again occupy the
coordination positions left vacant by O1 and O2. Note that,
although Glu-97 is shown in this diagram, it does not play a
direct role in this proposed mechanism of catalysis. Its role
appears to be to counteract the positive charge of the inferred
Fe^2+ ion and it does not mediate proton transfer.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
47261-47268)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Agarwal,
M.Rajavel,
B.Gopal,
and
N.Srinivasan
(2009).
Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold.
|
| |
PLoS One,
4,
e5736.
|
 |
|
|
|
|
 |
C.Roux,
N.Gresh,
L.E.Perera,
J.P.Piquemal,
and
L.Salmon
(2007).
Binding of 5-phospho-D-arabinonohydroxamate and 5-phospho-D-arabinonate inhibitors to zinc phosphomannose isomerase from Candida albicans studied by polarizable molecular mechanics and quantum mechanics.
|
| |
J Comput Chem,
28,
938-957.
|
 |
|
|
|
|
 |
M.Rhimi,
M.Juy,
N.Aghajari,
R.Haser,
and
S.Bejar
(2007).
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus stearothermophilus US100 L-arabinose isomerase by site-directed mutagenesis.
|
| |
J Bacteriol,
189,
3556-3563.
|
 |
|
|
|
|
 |
A.Teplyakov,
G.Obmolova,
J.Toedt,
M.Y.Galperin,
and
G.L.Gilliland
(2005).
Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism.
|
| |
J Bacteriol,
187,
5520-5527.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Siebers,
and
P.Schönheit
(2005).
Unusual pathways and enzymes of central carbohydrate metabolism in Archaea.
|
| |
Curr Opin Microbiol,
8,
695-705.
|
 |
|
|
|
|
 |
J.H.Lee,
and
C.J.Jeffery
(2005).
The crystal structure of rabbit phosphoglucose isomerase complexed with D-sorbitol-6-phosphate, an analog of the open chain form of D-glucose-6-phosphate.
|
| |
Protein Sci,
14,
727-734.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Hansen,
B.Schlichting,
J.Grötzinger,
M.K.Swan,
C.Davies,
and
P.Schönheit
(2005).
Mutagenesis of catalytically important residues of cupin type phosphoglucose isomerase from Archaeoglobus fulgidus.
|
| |
FEBS J,
272,
6266-6275.
|
 |
|
|
|
|
 |
T.Hansen,
B.Schlichting,
M.Felgendreher,
and
P.Schönheit
(2005).
Cupin-type phosphoglucose isomerases (Cupin-PGIs) constitute a novel metal-dependent PGI family representing a convergent line of PGI evolution.
|
| |
J Bacteriol,
187,
1621-1631.
|
 |
|
|
|
|
 |
T.Hansen,
and
P.Schönheit
(2005).
Escherichia coli phosphoglucose isomerase can be substituted by members of the PGI family, the PGI/PMI family, and the cPGI family.
|
| |
FEMS Microbiol Lett,
250,
49-53.
|
 |
|
|
|
|
 |
W.Hirano,
I.Gotoh,
T.Uekita,
and
M.Seiki
(2005).
Membrane-type 1 matrix metalloproteinase cytoplasmic tail binding protein-1 (MTCBP-1) acts as an eukaryotic aci-reductone dioxygenase (ARD) in the methionine salvage pathway.
|
| |
Genes Cells,
10,
565-574.
|
 |
|
|
|
|
 |
A.T.Cordeiro,
R.Hardré,
P.A.Michels,
L.Salmon,
L.F.Delboni,
and
O.H.Thiemann
(2004).
Leishmania mexicana mexicana glucose-6-phosphate isomerase: crystallization, molecular-replacement solution and inhibition.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
915-919.
|
 |
|
|
|
|
 |
H.Nishimasu,
S.Fushinobu,
H.Shoun,
and
T.Wakagi
(2004).
The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
|
| |
Structure,
12,
949-959.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

