 |
PDBsum entry 1qlt

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.3.38
- vanillyl-alcohol oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxy-3-methoxy-benzenemethanol + O2 = vanillin + H2O2
|
 |
 |
 |
 |
 |
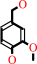 4-hydroxy-3-methoxy-benzenemethanol
4-hydroxy-3-methoxy-benzenemethanol
|
+
|
 O2
O2
|
=
|
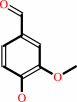 vanillin
vanillin
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
274:35514-35520
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase.
|
|
M.W.Fraaije,
R.H.van den Heuvel,
W.J.van Berkel,
A.Mattevi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
By mutating the target residue of covalent flavinylation in vanillyl-alcohol
oxidase, the functional role of the histidyl-FAD bond was studied. Three
His(422) mutants (H422A, H422T, and H422C) were purified, which all contained
tightly but noncovalently bound FAD. Steady state kinetics revealed that the
mutants have retained enzyme activity, although the turnover rates have
decreased by 1 order of magnitude. Stopped-flow analysis showed that the H422A
mutant is still able to form a stable binary complex of reduced enzyme and a
quinone methide product intermediate, a crucial step during vanillyl-alcohol
oxidase-mediated catalysis. The only significant change in the catalytic cycle
of the H422A mutant is a marked decrease in reduction rate. Redox potentials of
both wild type and H422A vanillyl-alcohol oxidase have been determined. During
reduction of H422A, a large portion of the neutral flavin semiquinone is
observed. Using suitable reference dyes, the redox potentials for the two
one-electron couples have been determined: -17 and -113 mV. Reduction of wild
type enzyme did not result in any formation of flavin semiquinone and revealed a
remarkably high redox potential of +55 mV. The marked decrease in redox
potential caused by the missing covalent histidyl-FAD bond is reflected in the
reduced rate of substrate-mediated flavin reduction limiting the turnover rate.
Elucidation of the crystal structure of the H422A mutant established that
deletion of the histidyl-FAD bond did not result in any significant structural
changes. These results clearly indicate that covalent interaction of the
isoalloxazine ring with the protein moiety can markedly increase the redox
potential of the flavin cofactor, thereby facilitating redox catalysis. Thus,
formation of a histidyl-FAD bond in specific flavoenzymes might have evolved as
a way to contribute to the enhancement of their oxidative power.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Ribbon representation of a vanillyl-alcohol
oxidase monomer. The histidyl-bound FAD cofactor is shown in a
ball-and-stick model. This figure was prepared with MOLSCRIPT
(55).
|
 |
Figure 5.
Fig. 5. Superposition of active site residues in the
unliganded H422A (shaded) and wild type VAO structures (black).
This figure was prepared with MOLSCRIPT (55).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(1999,
274,
35514-35520)
copyright 1999.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Fiorillo,
R.Federico,
F.Polticelli,
A.Boffi,
F.Mazzei,
M.Di Fusco,
A.Ilari,
and
P.Tavladoraki
(2011).
The structure of maize polyamine oxidase K300M mutant in complex with the natural substrates provides a snapshot of the catalytic mechanism of polyamine oxidation.
|
| |
FEBS J,
278,
809-821.
|
 |
|
|
|
|
 |
A.Winkler,
K.Motz,
S.Riedl,
M.Puhl,
P.Macheroux,
and
K.Gruber
(2009).
Structural and mechanistic studies reveal the functional role of bicovalent flavinylation in berberine bridge enzyme.
|
| |
J Biol Chem,
284,
19993-20001.
|
 |
|
|
|
|
 |
D.P.Heuts,
N.S.Scrutton,
W.S.McIntire,
and
M.W.Fraaije
(2009).
What's in a covalent bond? On the role and formation of covalently bound flavin cofactors.
|
| |
FEBS J,
276,
3405-3427.
|
 |
|
|
|
|
 |
O.Quaye,
S.Cowins,
and
G.Gadda
(2009).
Contribution of flavin covalent linkage with histidine 99 to the reaction catalyzed by choline oxidase.
|
| |
J Biol Chem,
284,
16990-16997.
|
 |
|
|
|
|
 |
C.H.Huang,
A.Winkler,
C.L.Chen,
W.L.Lai,
Y.C.Tsai,
P.Macheroux,
and
S.H.Liaw
(2008).
Functional roles of the 6-S-cysteinyl, 8alpha-N1-histidyl FAD in glucooligosaccharide oxidase from Acremonium strictum.
|
| |
J Biol Chem,
283,
30990-30996.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.W.van Hellemond,
H.Mazon,
A.J.Heck,
R.H.van den Heuvel,
D.P.Heuts,
D.B.Janssen,
and
M.W.Fraaije
(2008).
ADP competes with FAD binding in putrescine oxidase.
|
| |
J Biol Chem,
283,
28259-28264.
|
 |
|
|
|
|
 |
J.Jin,
H.Mazon,
R.H.van den Heuvel,
A.J.Heck,
D.B.Janssen,
and
M.W.Fraaije
(2008).
Covalent flavinylation of vanillyl-alcohol oxidase is an autocatalytic process.
|
| |
FEBS J,
275,
5191-5200.
|
 |
|
|
|
|
 |
N.G.Leferink,
W.A.van den Berg,
and
W.J.van Berkel
(2008).
l-Galactono-gamma-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis.
|
| |
FEBS J,
275,
713-726.
|
 |
|
|
|
|
 |
M.Kujawa,
J.Volc,
P.Halada,
P.Sedmera,
C.Divne,
C.Sygmund,
C.Leitner,
C.Peterbauer,
and
D.Haltrich
(2007).
Properties of pyranose dehydrogenase purified from the litter-degrading fungus Agaricus xanthoderma.
|
| |
FEBS J,
274,
879-894.
|
 |
|
|
|
|
 |
T.Ohta,
T.Kawabata,
K.Nishikawa,
A.Tani,
K.Kimbara,
and
F.Kawai
(2006).
Analysis of amino acid residues involved in catalysis of polyethylene glycol dehydrogenase from Sphingopyxis terrae, using three-dimensional molecular modeling-based kinetic characterization of mutants.
|
| |
Appl Environ Microbiol,
72,
4388-4396.
|
 |
|
|
|
|
 |
M.H.Lee,
W.L.Lai,
S.F.Lin,
C.S.Hsu,
S.H.Liaw,
and
Y.C.Tsai
(2005).
Structural characterization of glucooligosaccharide oxidase from Acremonium strictum.
|
| |
Appl Environ Microbiol,
71,
8881-8887.
|
 |
|
|
|
|
 |
S.R.Wilkinson,
S.R.Prathalingam,
M.C.Taylor,
D.Horn,
and
J.M.Kelly
(2005).
Vitamin C biosynthesis in trypanosomes: a role for the glycosome.
|
| |
Proc Natl Acad Sci U S A,
102,
11645-11650.
|
 |
|
|
|
|
 |
C.B.Chiribau,
C.Sandu,
M.Fraaije,
E.Schiltz,
and
R.Brandsch
(2004).
A novel gamma-N-methylaminobutyrate demethylating oxidase involved in catabolism of the tobacco alkaloid nicotine by Arthrobacter nicotinovorans pAO1.
|
| |
Eur J Biochem,
271,
4677-4684.
|
 |
|
|
|
|
 |
H.Safi,
P.F.Barnes,
D.L.Lakey,
H.Shams,
B.Samten,
R.Vankayalapati,
and
S.T.Howard
(2004).
IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis.
|
| |
Mol Microbiol,
52,
999.
|
 |
|
|
|
|
 |
M.H.Hefti,
J.Vervoort,
and
W.J.van Berkel
(2003).
Deflavination and reconstitution of flavoproteins.
|
| |
Eur J Biochem,
270,
4227-4242.
|
 |
|
|
|
|
 |
D.E.Edmondson,
and
P.Newton-Vinson
(2001).
The covalent FAD of monoamine oxidase: structural and functional role and mechanism of the flavinylation reaction.
|
| |
Antioxid Redox Signal,
3,
789-806.
|
 |
|
|
|
|
 |
I.Efimov,
C.N.Cronin,
and
W.S.McIntire
(2001).
Effects of noncovalent and covalent FAD binding on the redox and catalytic properties of p-cresol methylhydroxylase.
|
| |
Biochemistry,
40,
2155-2166.
|
 |
|
|
|
|
 |
M.Eschenbrenner,
L.J.Chlumsky,
P.Khanna,
F.Strasser,
and
M.S.Jorns
(2001).
Organization of the multiple coenzymes and subunits and role of the covalent flavin link in the complex heterotetrameric sarcosine oxidase.
|
| |
Biochemistry,
40,
5352-5367.
|
 |
|
|
|
|
 |
N.Tahallah,
M.Pinkse,
C.S.Maier,
and
A.J.Heck
(2001).
The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument.
|
| |
Rapid Commun Mass Spectrom,
15,
596-601.
|
 |
|
|
|
|
 |
A.Albert,
M.Martínez-Ripoll,
A.Espinosa-Ruiz,
L.Yenush,
F.A.Culiáñez-Macià,
and
R.Serrano
(2000).
The X-ray structure of the FMN-binding protein AtHal3 provides the structural basis for the activity of a regulatory subunit involved in signal transduction.
|
| |
Structure,
8,
961-969.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.H.van Den Heuvel,
M.W.Fraaije,
M.Ferrer,
A.Mattevi,
and
W.J.van Berkel
(2000).
Inversion of stereospecificity of vanillyl-alcohol oxidase.
|
| |
Proc Natl Acad Sci U S A,
97,
9455-9460.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

