 |
PDBsum entry 1ql1
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
56:137-150
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
The molecular structure and structural transition of the alpha-helical capsid in filamentous bacteriophage Pf1.
|
|
L.C.Welsh,
M.F.Symmons,
D.A.Marvin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The major coat protein in the capsid of Pf1 filamentous bacteriophage (Inovirus)
forms a helical assembly of about 7000 identical protein subunits, each of which
contains 46 amino-acid residues and can be closely approximated by a single
gently curved alpha-helix. Since the viral DNA occupies the core of the tubular
capsid and appears to make no significant specific interactions with the capsid
proteins, the capsid is a simple model system for the study of the static and
dynamic properties of alpha-helix assembly. The capsid undergoes a reversible
temperature-induced structural transition at about 283 K between two slightly
different helix forms. The two forms can coexist without an intermediate state,
consistent with a first-order structural phase transition. The molecular model
of the higher temperature form was refined using improved X-ray fibre
diffraction data and new refinement and validation methods. The refinement
indicates that the two forms are related by a change in the orientation of the
capsid subunits within the virion, without a significant change in local
conformation of the subunits. On the higher temperature diffraction pattern
there is a region of observed intensity that is not consistent with a simple
helix of identical subunits; it is proposed that the structure involves groups
of three subunits which each have a slightly different orientation within the
group. The grouping of subunits suggests that a change in subunit libration
frequency could be the basis of the Pf1 structural transition; calculations from
the model are used to explore this idea.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 The Pf1^L subunit 4ifm (solid lines) and the Pf1^H
subunit 2ifn (broken lines). The relative position of model 4ifm
was altered by rotating and translating the coordinates with
respect to the virion axis in order to superimpose the centres
of the subunits. Lines connect C^  atoms.
Steroview from outside the virion towards the virion axis, which
is shown as a vertical line. atoms.
Steroview from outside the virion towards the virion axis, which
is shown as a vertical line.
|
 |
Figure 8.
Figure 8 Grouping of subunits in the perturbed helix. Views
perpendicular to the virion axis, N-termini of the subunits
towards the top. Colour coding as in Fig. 6-. (a) Units k = 1
and k = 2 in the virion basic helix are superposed on unit k = 0
by the operation (-kTH, -kHH). Black, model RPf1^H; red, unit k
= 0 of model 3RPf1^H; green, k = 1; blue, k = 2. Heavy lines
connect C^  atoms;
lighter lines connect side-chain non-H atoms. Stereoview from
outside the virion towards the virion axis, which is shown as a
vertical line. (b) Models as (a) but viewed at 90° to (a),
tangent to the circumference of a cylinder coaxial with the
virion, in the direction of increasing cylindrical polar angle atoms;
lighter lines connect side-chain non-H atoms. Stereoview from
outside the virion towards the virion axis, which is shown as a
vertical line. (b) Models as (a) but viewed at 90° to (a),
tangent to the circumference of a cylinder coaxial with the
virion, in the direction of increasing cylindrical polar angle
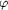 .
(c) The assembly of subunits in model 3RPf1^H. Each subunit is
represented as a space-filling coil following the protein
backbone at 5 Å radius. Axial slab about 100 Å long,
corresponding to about 0.5% of the total length of the virion.
Colour coding as in (a): red, k = 0, 3, 6, 9, ...; green, k = 1,
4, 7, 10, ...; blue, k = 2, 5, 8, 11, ... . Three adjacent
subunits (k = 0, -6, -11) are shown in atomic detail (white
lines) within `transparent' rods. Heavy lines connect C^ .
(c) The assembly of subunits in model 3RPf1^H. Each subunit is
represented as a space-filling coil following the protein
backbone at 5 Å radius. Axial slab about 100 Å long,
corresponding to about 0.5% of the total length of the virion.
Colour coding as in (a): red, k = 0, 3, 6, 9, ...; green, k = 1,
4, 7, 10, ...; blue, k = 2, 5, 8, 11, ... . Three adjacent
subunits (k = 0, -6, -11) are shown in atomic detail (white
lines) within `transparent' rods. Heavy lines connect C^  atoms;
lighter lines connect side-chain non-H atoms. atoms;
lighter lines connect side-chain non-H atoms.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2000,
56,
137-150)
copyright 2000.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.K.Straus,
W.R.Scott,
C.D.Schwieters,
and
D.A.Marvin
(2011).
Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR.
|
| |
Eur Biophys J,
40,
221-234.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Tsuboi,
M.Tsunoda,
S.A.Overman,
J.M.Benevides,
and
G.J.Thomas
(2010).
A structural model for the single-stranded DNA genome of filamentous bacteriophage Pf1.
|
| |
Biochemistry,
49,
1737-1743.
|
 |
|
|
|
|
 |
J.L.Lorieau,
L.A.Day,
and
A.E.McDermott
(2008).
Conformational dynamics of an intact virus: order parameters for the coat protein of Pf1 bacteriophage.
|
| |
Proc Natl Acad Sci U S A,
105,
10366-10371.
|
 |
|
|
|
|
 |
S.J.Opella,
A.C.Zeri,
and
S.H.Park
(2008).
Structure, dynamics, and assembly of filamentous bacteriophages by nuclear magnetic resonance spectroscopy.
|
| |
Annu Rev Phys Chem,
59,
635-657.
|
 |
|
|
|
|
 |
S.K.Straus,
W.R.Scott,
and
D.A.Marvin
(2008).
The hand of the filamentous bacteriophage helix.
|
| |
Eur Biophys J,
37,
1077-1082.
|
 |
|
|
|
|
 |
S.K.Straus,
W.R.Scott,
M.F.Symmons,
and
D.A.Marvin
(2008).
On the structures of filamentous bacteriophage Ff (fd, f1, M13).
|
| |
Eur Biophys J,
37,
521-527.
|
 |
|
|
|
|
 |
D.S.Thiriot,
A.A.Nevzorov,
and
S.J.Opella
(2005).
Structural basis of the temperature transition of Pf1 bacteriophage.
|
| |
Protein Sci,
14,
1064-1070.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Tsuboi,
S.A.Overman,
K.Nakamura,
A.Rodriguez-Casado,
and
G.J.Thomas
(2003).
Orientation and interactions of an essential tryptophan (Trp-38) in the capsid subunit of Pf3 filamentous virus.
|
| |
Biophys J,
84,
1969-1976.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

